RNA binding proteins (RBPs) are a large protein family that plays roles at all level of gene regulation through interacting with RNAs, and are required for all biological processes. EuRBPDB is a comprehensive and user-friendly database of eukaryotic RBPs. It classifies and annotates RBPs in 162 eukaryotic genomes. EuRBPDB totally contains 315,222 RBPs, which are further classified into 791 families based on their RNA-binding domain (RBD). EuRBPDB provides a platform to connect RBPs with multi-layer information of their characteristics and function, including RNA binding and gene transcription landscape. Since many RBPs have been found to be involved in the regulation of progression of cancer, EuRBPDB collected the cancer associated information of RBPs from literatures, TCGA, ENCODE and LINC project, and found 308 have been reported to be cancer relevant. Moreover, our analysis also revealed that 637 RBPs, which have not yet been reported in any literature, might regulate the progression of cancer. All cancer associated RBPs regardless of whether they have been published, are described in detailed in EuRBPDB. EuRBPDB is helpful for biologists to generate novel hypotheses about the roles and regulatory mechanisms of RBPs in various physiological and cancer processes.
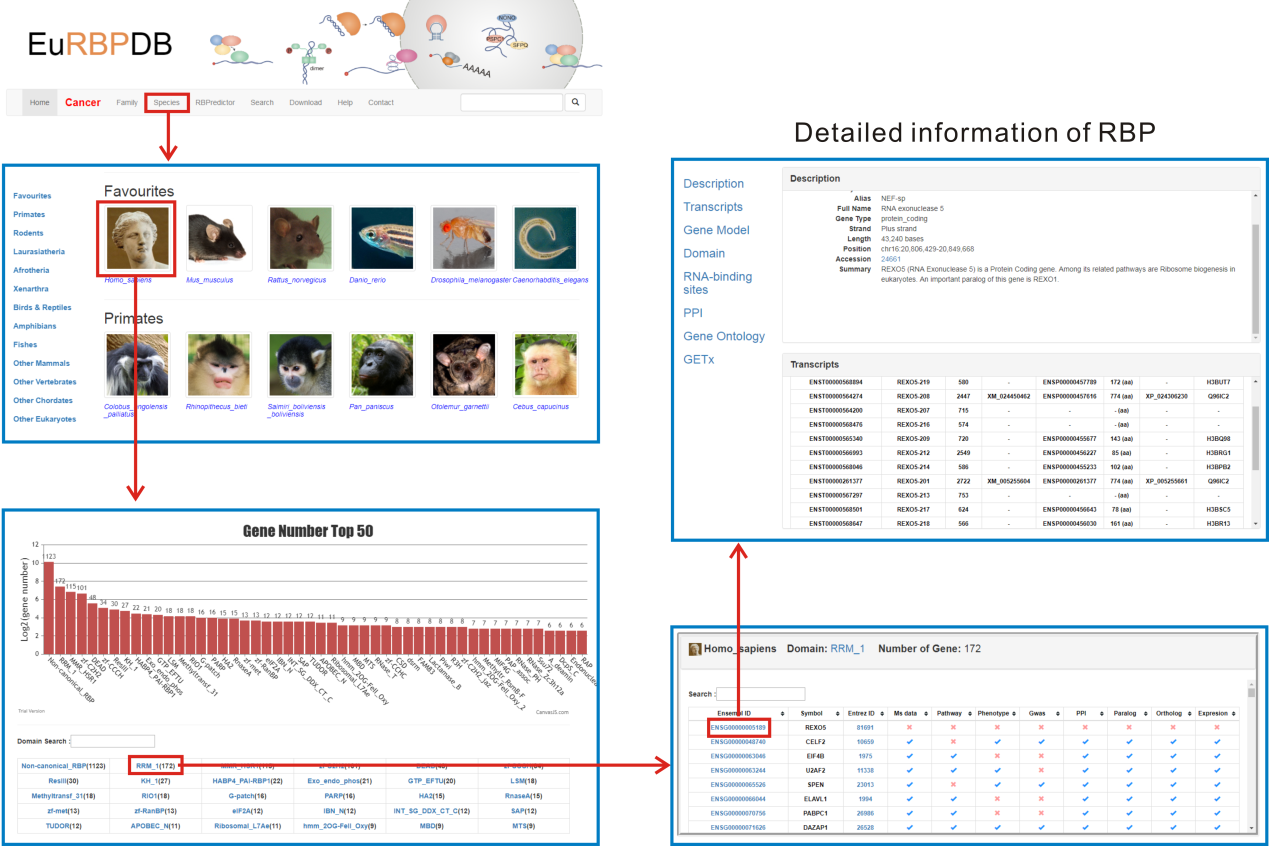
1. Browse by species. On the "Species" page, 162 species were classified into 12 categories according to Ensembl taxonomy. To make it convenient for users to browse, we put a favourites category on the top of species page. Users can browse the database by clicking the species image of interest, and retrieve the detailed RBP information through the following steps: families->family gene list ->single gene annotation
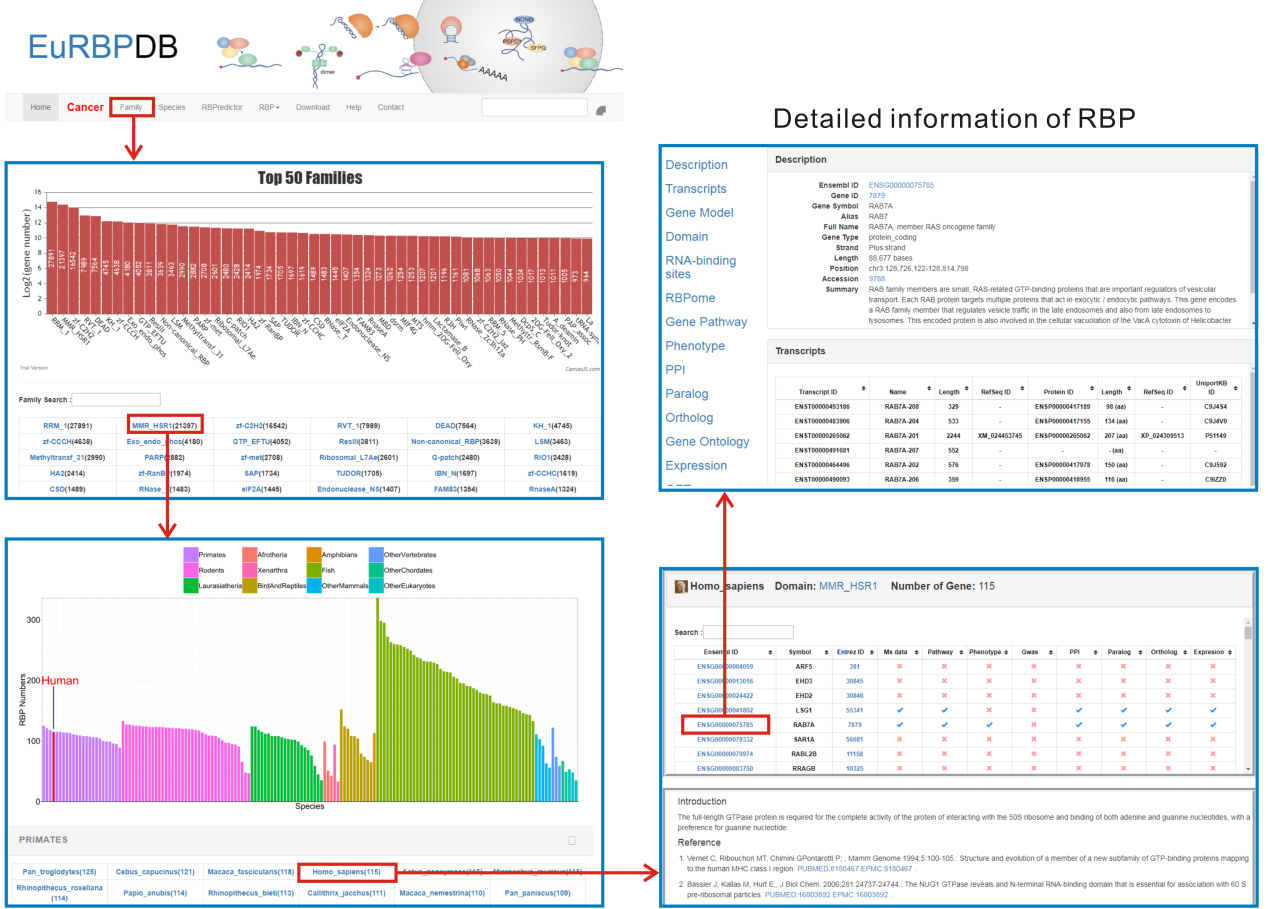
2. Browse by family. On the "Family" page, EuRBPDB lists all 686 RBP families from 162 eukaryotes. RBP families were ordered by family size in descending order. By clicking the family name, users will get all RBPs grouped by species in this family. Users can browse the database by click the family name, and obtain the detailed information of RBP through the following steps: species->gene list ->single gene annotation.
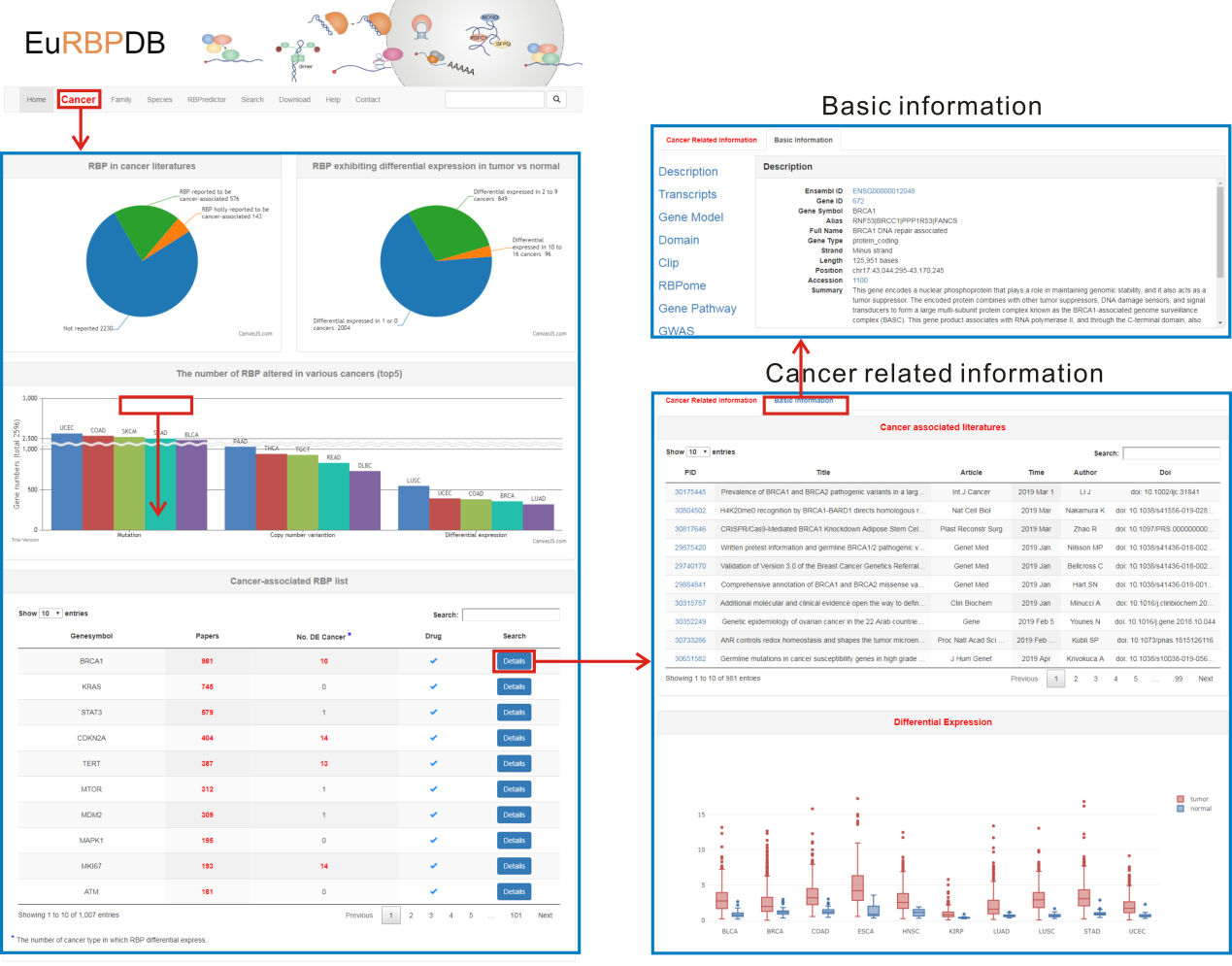
3. Browse by cancer. On the "Cancer" page, EuRBPDB lists all 1,364 cancer-associated RBPs from human. By click the "Details" link, users can obtain the detailed annotation information of RBPs. EuRBPDB provides the overview of the cancer-associated RBPs in "Cancer" page, showing how many RBPs were reported to be cancer-associated, the number of RBP differentially expressed in cancers, and the top 5 cancers with most mutated, copy number variation (CNV) and differentially expressed RBPs.
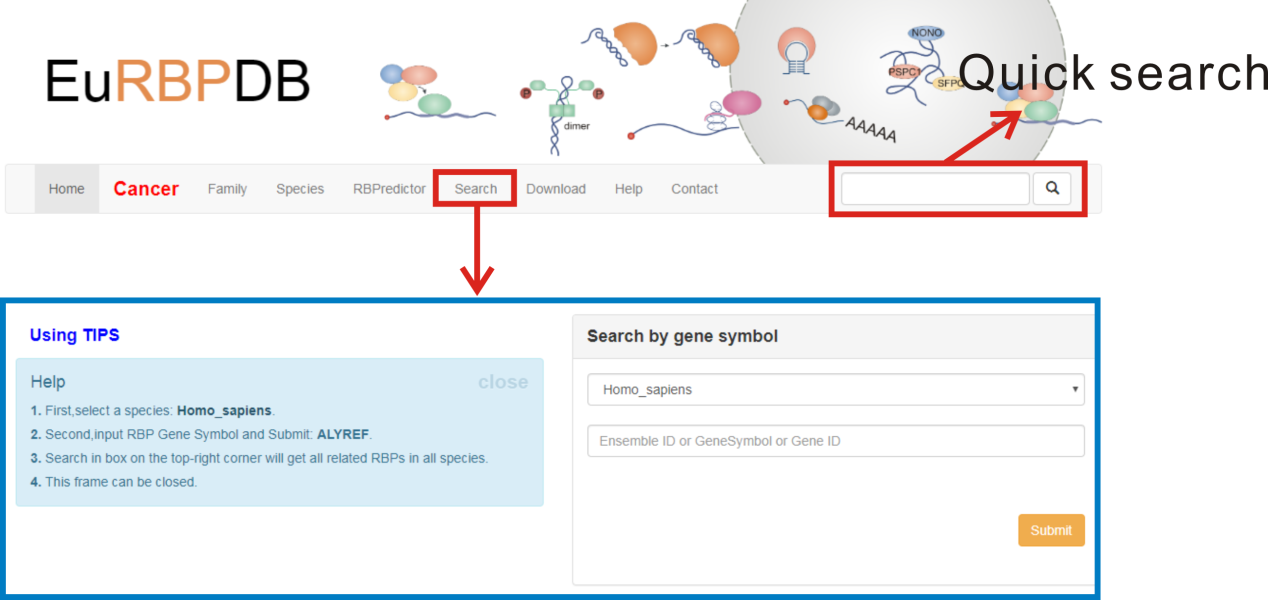
1. A quick search box for Ensembl gene id or gene symbol locates at the right corner of navigation bar, and the search will return all RBPs in any species matching the searching criteria.
2. To browse the detailed information of any specific RBP, users can specify both the species and RBP name/ID in "Search" page.
To help users identify RBPs from their own protein sequences, we set up a RBP prediction server, RBPredictor. Such RBP prediction is based on the RBD sets used in this study, and we performed hmm-search program in HMMER (v3.2.1) package to determine whether the protein sequence submitted is a putative RBP. In "RBPredictor" page, users is only required to input one or multiple protein sequences in fasta format, or submit a fasta file with protein sequences. Currently, users can upload up to 1000 protein sequences in one time and obtain results within a few minutes. If an input protein is identified as a putative RBP, RBPredictor will also list all potential RBDs.
The gene lists and protein sequences of RBPs for each species, as well as all RBD HMM profiles could be downloaded from the download page.
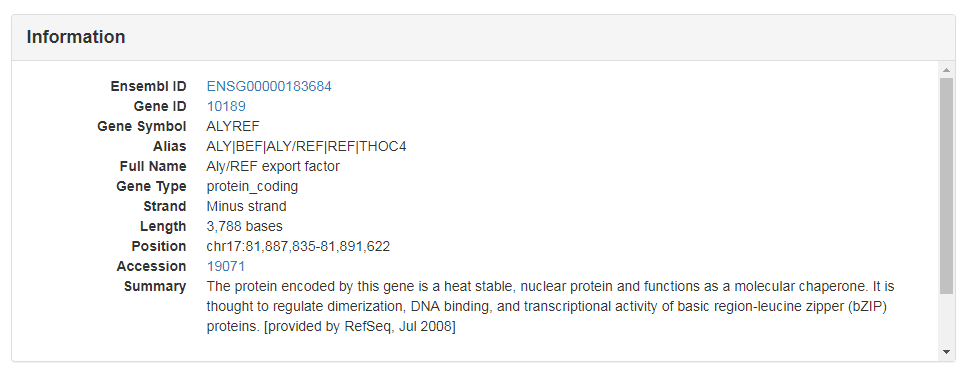
This section shows the Ensembl ID, Gene ID, Gene Symbol, Alias, Full Name, Gene Type, Strand, Length, Position and Transcripts information which were extracted from Ensembl or GeneCards database.
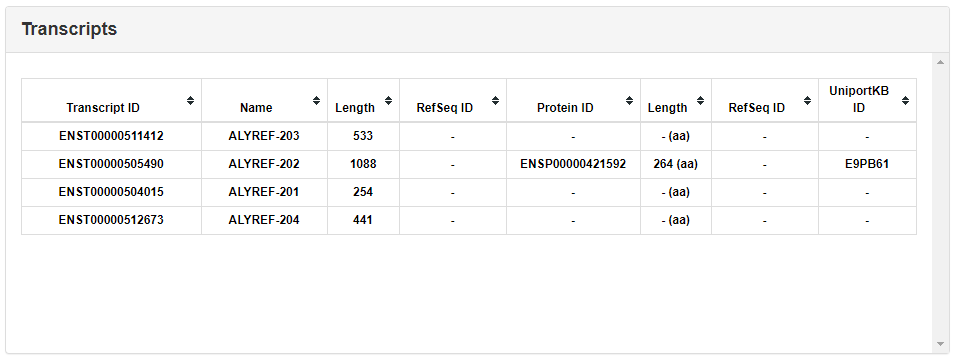
This section lists all isoforms of a RBP gene. Users can obtain Ensembl transcript ID, Name, length RefSeq ID, Ensembl protein ID, protein length, and UniportKB ID of each isoform from this section.
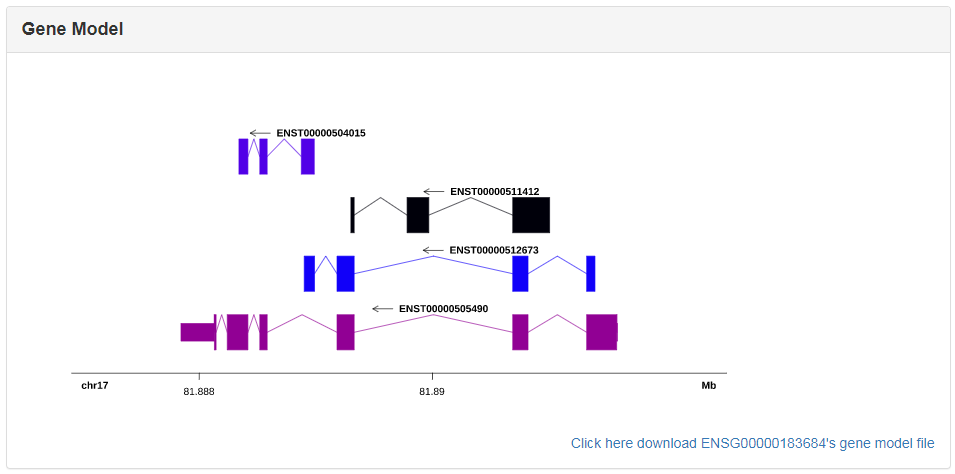
This section shows the distribution of the CDS, UTR and intron of a gene on chromosome based on the information from Ensembl gtf files. The high resolution gene model figure can be downloaded conveniently by clicking the lower right corner link.
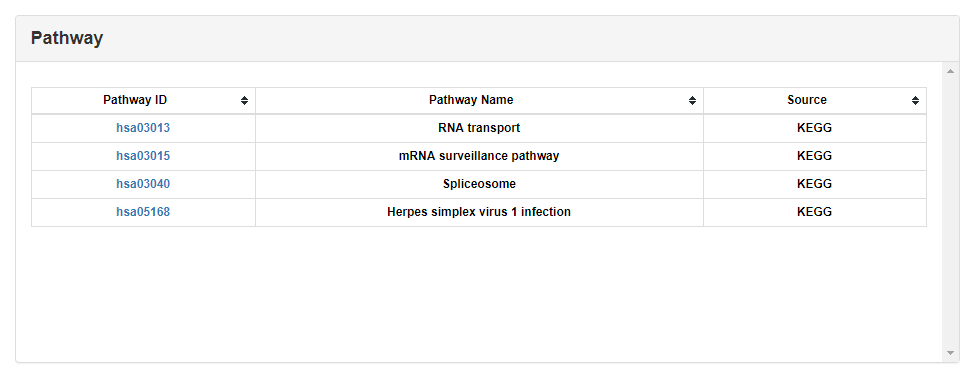
Detailed information of all RBP domains of RBPs found by hmmsearcher is shown in this part.
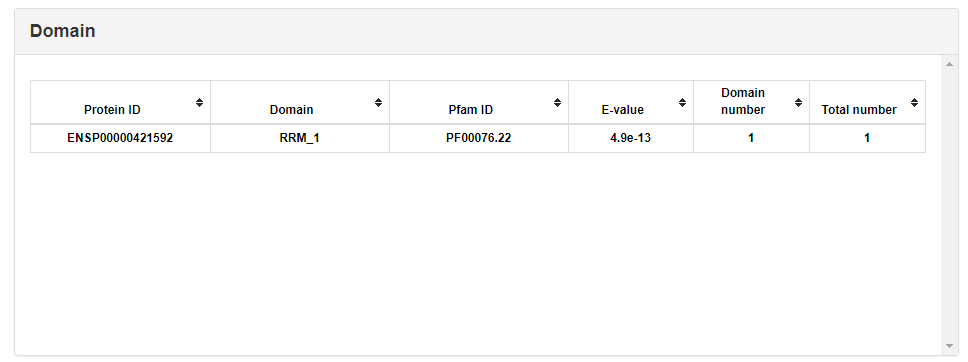
The protein-protein interactions were extracted by STRING API. The detailed interaction information can be downloaded by clicking the lower right corner link.
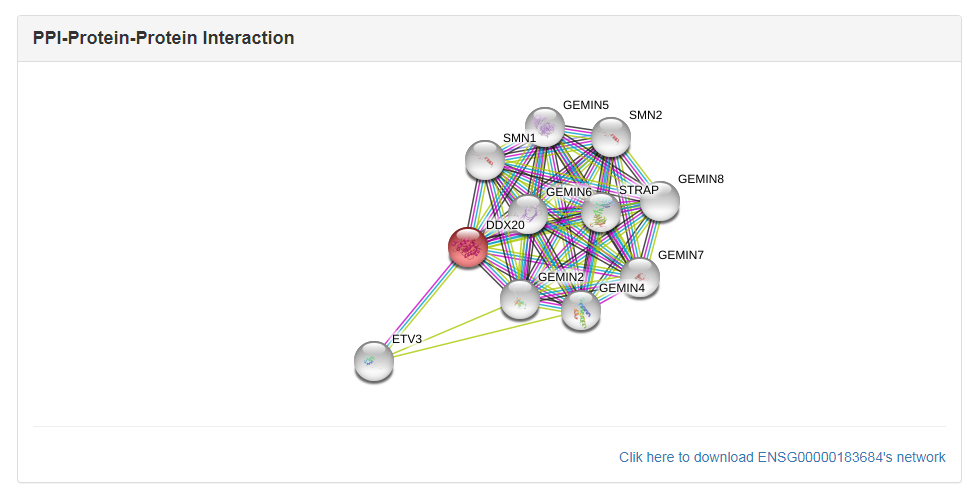
The GO annotations were parsed from gene2go file, which was downloaded from NCBI ftp.
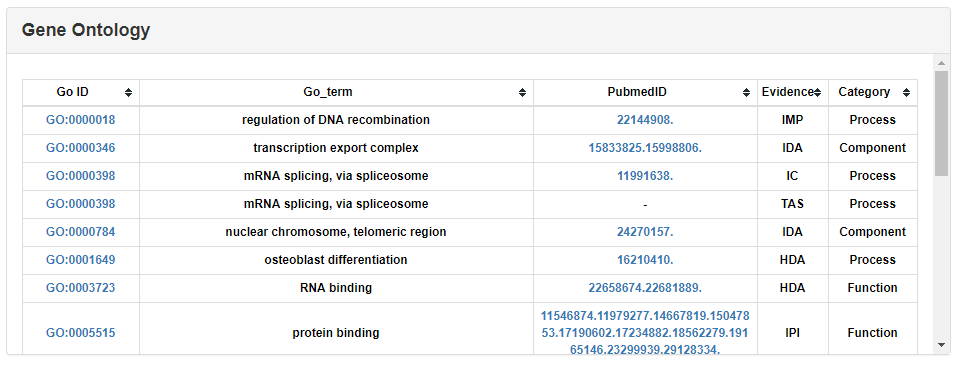
The Pathway annotations were downloaded from KEGG database.
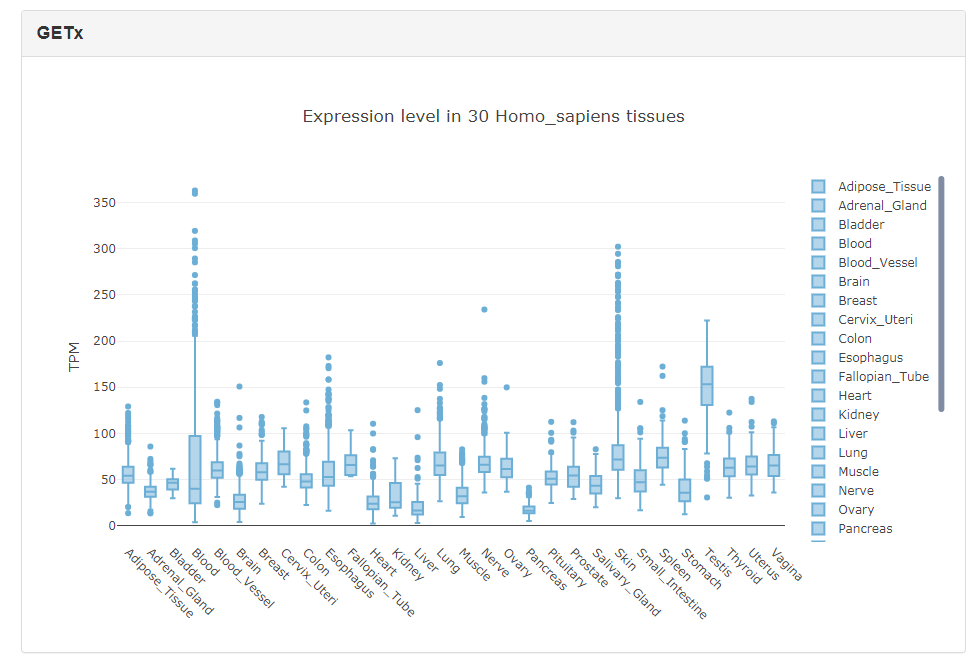
The expression levels of RBPs in different tissues were obtained from public databases such as GTEx. Currently, EuRBPDB only contains RBP expression information in human, mouse and rat. The expression level of each RBP is shown in boxplot as the following figure, and users can add or remove sample from the boxplot through clicking the sample name in the right panel.
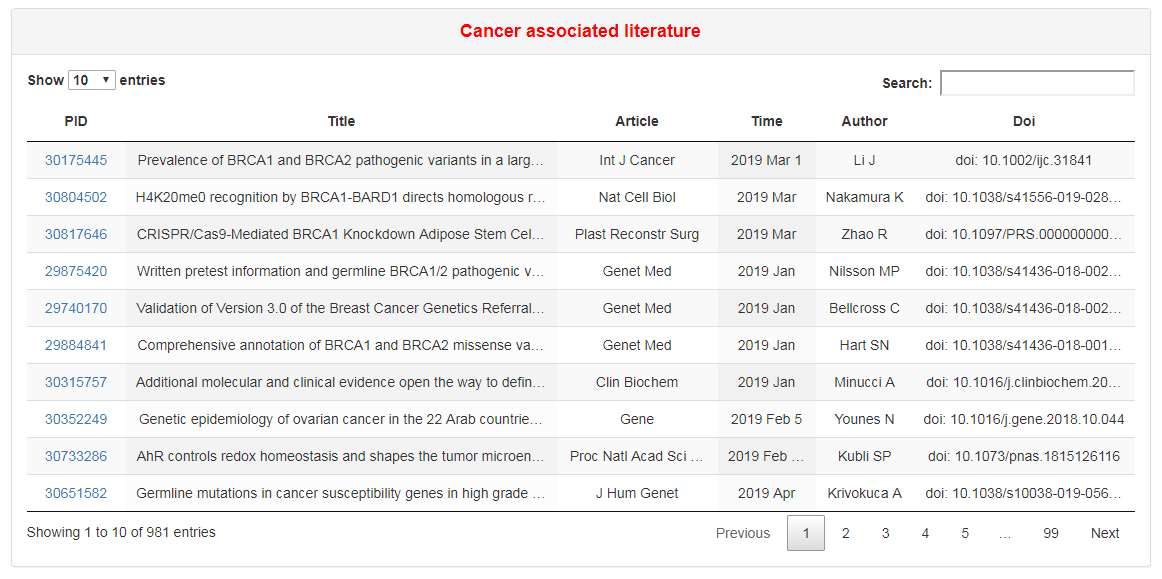
This part lists all cancer-associated literatures of a RBP. These literatures were found by geneclip3 server (http://ci.smu.edu.cn/genclip3/).
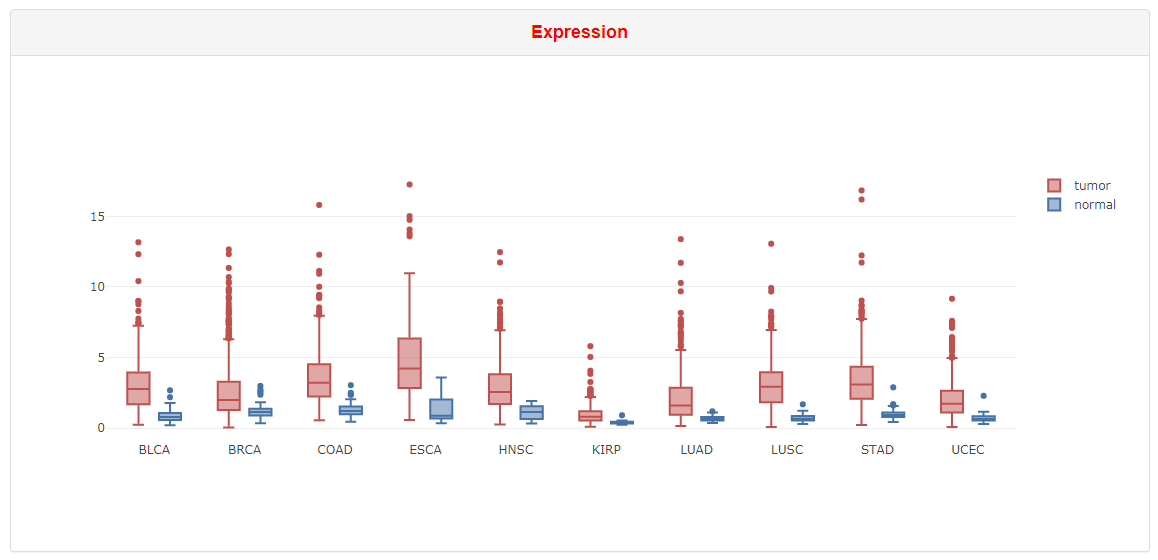
The boxplot shows the expression level of a RBP in tumor and normal tissues. Only cancers exhibiting differential expression of selected RBP were shown in the boxplot.
This part shows the expression level of an RBP in 33 cancers. Users can add or remove sample from the boxplot through clicking the sample name in the right panel.

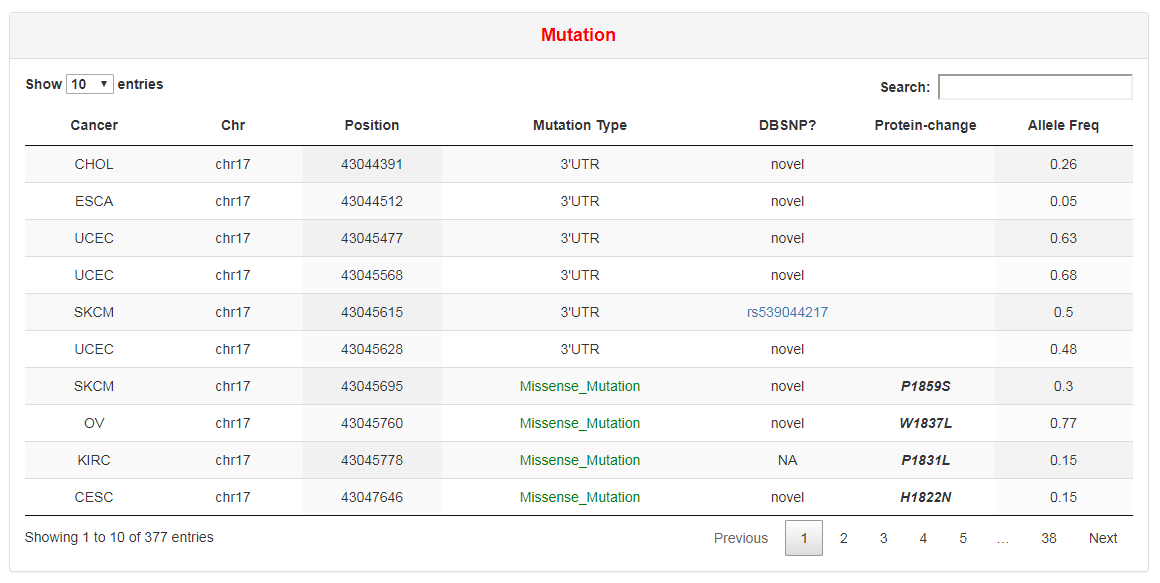
The table lists all mutations of a RBP. Users can obtain the mutation type, genomic position, SNP ID in dbSNP database, amino acid changes and mutation frequency of each mutation in each cancer from the table.
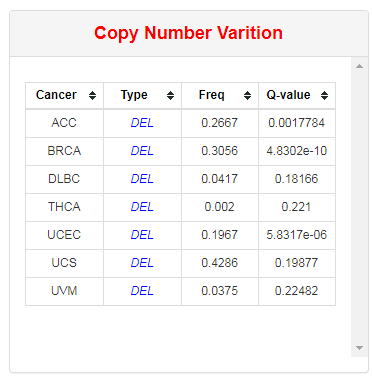
This part lists all cancers with deletion or amplification of selected RBP.
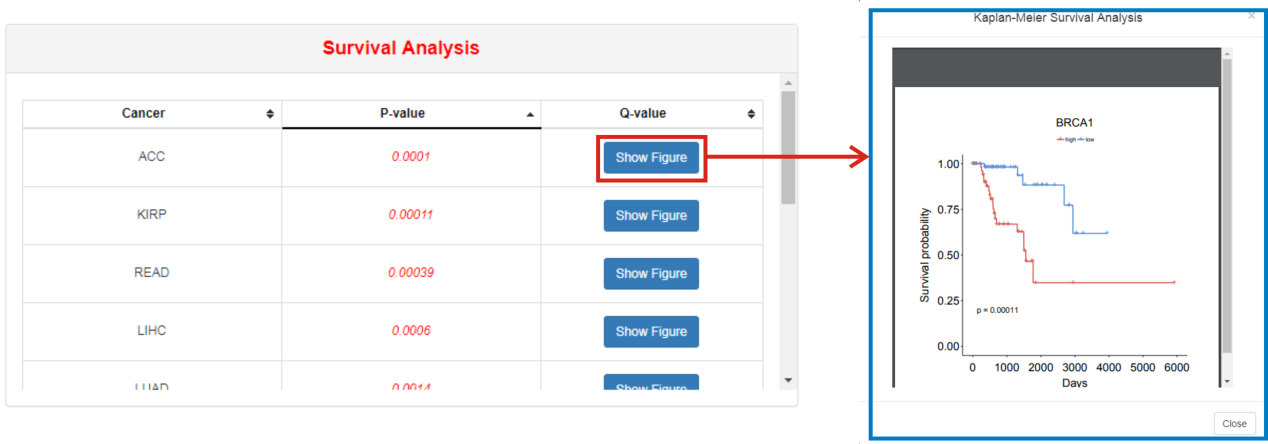
This part lists all cancers with significantly different survival state between high expression group and low expression group of selected RBP. Clicking "Show Figure" will generate a Kaplan-Meier survival plot which can be downloaded as PDF format.
This part contains data from two L1000 assay level-5 datasets (GSE92742 and GSE70138) (26) generated by the Library of Integrated Cellular Signatures (LINCS) project which were downloaded from GEO. These datasets contain over 1,600,000 subdatasets measuring the effects 30,744 drugs on the RNA profiles of 44 cell lines. In this section, users can obtain the expression alteration of the selected RBP simply by entering the drug name into "Input Drug" box and cell line name into "Input Cell Line" box, and then clicking the submit button. The website will return the z-score boxplot of selected RBP. Drug and cell line list can be found in the "Cell lines and drugs in GSE70138" and "Cell lines and drugs in GSE92742" links.

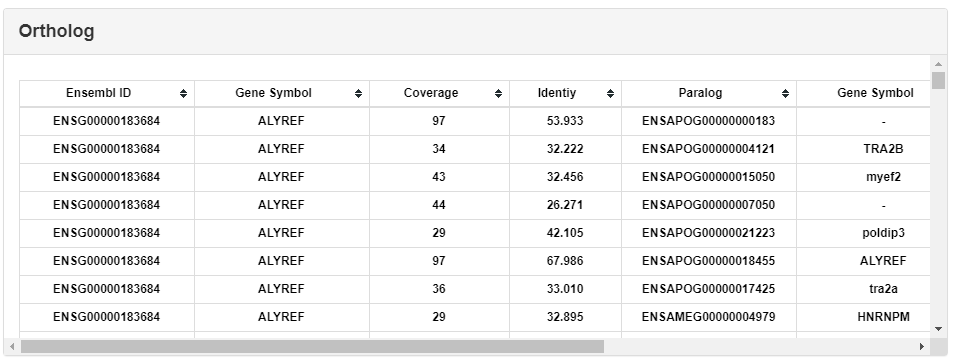
The reciprocal best hit (RBH) method (22) was used to predict the putative orthologs of RBPs among different species. We have performed all-against-all BLASTP (v2.7.1+) search between proteins of two genomes with strict cutoffs (E-value ≤ 1e-6, coverage ≥ 50%, identity ≥ 30%) and annotated the reciprocal best hit pairs as orthologs. This part lists all paralogs of the selected RBP.
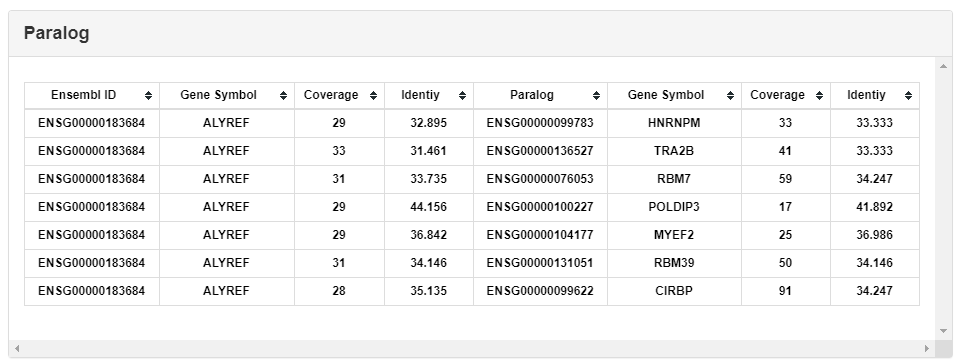
Paralogs was predicted by the BLAST score ratio (BSR) (23) approach. BLASTP search has been conducted in each genome with the same parameters as in orthologs search. The BSR value cutoff was set to 0.4. This part lists all orthologs of selected RBPs.