
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| COAD | |||
| DLBC | |||
| LUSC | |||
| OV | |||
| PAAD | |||
| READ | |||
| TGCT | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| LUSC | ||
| ESCA | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| PCPG | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| LUAD |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 7893829 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000630279 | FKBP4-208 | 156 | - | ENSP00000486800 | 51 (aa) | - | F5H120 |
| ENST00000539181 | FKBP4-203 | 370 | - | ENSP00000438491 | 84 (aa) | - | H0YFG2 |
| ENST00000540260 | FKBP4-204 | 777 | - | - | - (aa) | - | - |
| ENST00000543769 | FKBP4-206 | 425 | - | ENSP00000439703 | 51 (aa) | - | F5H120 |
| ENST00000001008 | FKBP4-201 | 3732 | XM_011520929 | ENSP00000001008 | 459 (aa) | XP_011519231 | Q02790 |
| ENST00000538622 | FKBP4-202 | 572 | - | ENSP00000446368 | 86 (aa) | - | F5H1U3 |
| ENST00000543037 | FKBP4-205 | 727 | - | - | - (aa) | - | - |
| ENST00000544366 | FKBP4-207 | 287 | - | ENSP00000442193 | 23 (aa) | - | H0YG86 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04915 | Estrogen signaling pathway | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000004478 | rs56196860 | 12 | 2799164 | ? | Heel bone mineral density | 30048462 | [0.024-0.047] unit decrease | 0.0354551 | EFO_0009270 |
| ENSG00000004478 | rs56196860 | 12 | 2799164 | ? | Lung function (FVC) | 30595370 | EFO_0004312 | ||
| ENSG00000004478 | rs56196860 | 12 | 2799164 | ? | Waist-hip ratio | 30595370 | EFO_0004343 | ||
| ENSG00000004478 | rs56196860 | 12 | 2799164 | A | Lung function (FVC) | 30804560 | [0.04-0.067] unit decrease | 0.0531 | EFO_0004312 |
| ENSG00000004478 | rs56196860 | 12 | 2799164 | A | FEV1 | 30804560 | [0.031-0.058] unit decrease | 0.0446 | EFO_0004314 |
| ENSG00000004478 | rs56196860 | 12 | 2799164 | A | Peak expiratory flow | 30804560 | [0.02-0.049] unit decrease | 0.0345 | |
| ENSG00000004478 | rs56196860 | 12 | 2799164 | ? | Heel bone mineral density | 30595370 | EFO_0009270 |
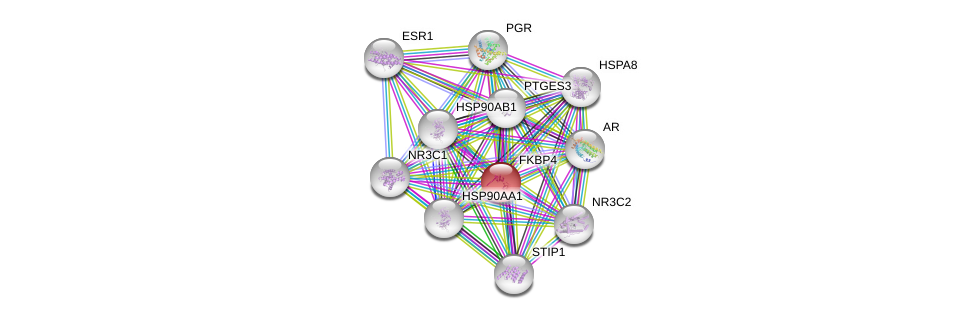
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000413 | protein peptidyl-prolyl isomerization | 21873635. | IBA | Process |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | 21873635. | IBA | Function |
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | 11350175. | IDA | Function |
| GO:0005515 | protein binding | 11583998.12604780.14638689.19875381.20188096.21170051.21360678.21730179.23741051.25036637.25277244. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005528 | FK506 binding | 1376003. | TAS | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005739 | mitochondrion | - | IEA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | ISS | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005874 | microtubule | - | IEA | Component |
| GO:0006457 | protein folding | 1279700. | TAS | Process |
| GO:0006463 | steroid hormone receptor complex assembly | - | IEA | Process |
| GO:0006825 | copper ion transport | - | IEA | Process |
| GO:0007566 | embryo implantation | - | IEA | Process |
| GO:0010977 | negative regulation of neuron projection development | - | ISS | Process |
| GO:0030521 | androgen receptor signaling pathway | - | IEA | Process |
| GO:0030674 | protein binding, bridging | 2378870. | TAS | Function |
| GO:0030850 | prostate gland development | - | IEA | Process |
| GO:0031072 | heat shock protein binding | 9660753. | IPI | Function |
| GO:0031111 | negative regulation of microtubule polymerization or depolymerization | - | ISS | Process |
| GO:0031115 | negative regulation of microtubule polymerization | - | IEA | Process |
| GO:0031503 | protein-containing complex localization | - | IEA | Process |
| GO:0032767 | copper-dependent protein binding | - | IEA | Function |
| GO:0032991 | protein-containing complex | - | IEA | Component |
| GO:0035259 | glucocorticoid receptor binding | - | IEA | Function |
| GO:0043025 | neuronal cell body | - | IEA | Component |
| GO:0044295 | axonal growth cone | - | ISS | Component |
| GO:0046661 | male sex differentiation | - | IEA | Process |
| GO:0048156 | tau protein binding | - | IEA | Function |
| GO:0048471 | perinuclear region of cytoplasm | - | IEA | Component |
| GO:0051219 | phosphoprotein binding | - | IEA | Function |
| GO:0061077 | chaperone-mediated protein folding | 21873635. | IBA | Process |
| GO:0061077 | chaperone-mediated protein folding | 11350175. | IDA | Process |
| GO:0070062 | extracellular exosome | 19056867.20458337. | HDA | Component |
| GO:1900034 | regulation of cellular response to heat | - | TAS | Process |