
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRC | |||
| TGCT | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| SARC | ||
| HNSC | ||
| SKCM | ||
| BRCA | ||
| ESCA | ||
| COAD | ||
| CESC | ||
| UCEC | ||
| GBM | ||
| LIHC | ||
| CHOL | ||
| THCA |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000475354 | SLC25A5-204 | 483 | - | - | - (aa) | - | - |
| ENST00000463551 | SLC25A5-203 | 433 | - | - | - (aa) | - | - |
| ENST00000460013 | SLC25A5-202 | 775 | - | - | - (aa) | - | - |
| ENST00000317881 | SLC25A5-201 | 1274 | - | ENSP00000360671 | 298 (aa) | - | P05141 |
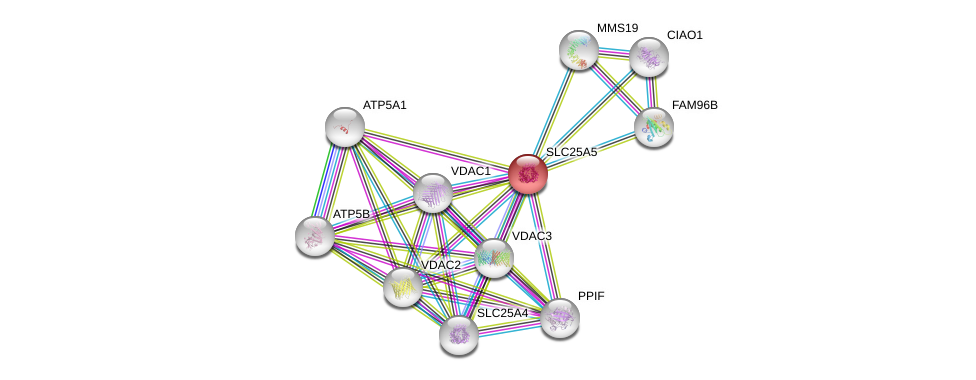
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000005022 | SLC25A5 | 100 | 98.322 | ENSMUSG00000016319 | Slc25a5 | 100 | 98.322 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0005471 | ATP:ADP antiporter activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 17715127.19116139.19154410.21370995.24725412. | IPI | Function |
| GO:0005634 | nucleus | 21630459. | HDA | Component |
| GO:0005739 | mitochondrion | 20833797. | HDA | Component |
| GO:0005739 | mitochondrion | - | IDA | Component |
| GO:0005743 | mitochondrial inner membrane | - | TAS | Component |
| GO:0005887 | integral component of plasma membrane | 2168878. | TAS | Component |
| GO:0007059 | chromosome segregation | - | IEA | Process |
| GO:0008284 | positive regulation of cell proliferation | 19154410. | IMP | Process |
| GO:0015207 | adenine transmembrane transporter activity | 2168878. | TAS | Function |
| GO:0015853 | adenine transport | - | IEA | Process |
| GO:0015866 | ADP transport | - | IEA | Process |
| GO:0015867 | ATP transport | - | IEA | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0031625 | ubiquitin protein ligase binding | 19725078. | IPI | Function |
| GO:0042645 | mitochondrial nucleoid | 18063578. | IDA | Component |
| GO:0045121 | membrane raft | - | IEA | Component |
| GO:0050796 | regulation of insulin secretion | - | TAS | Process |
| GO:0055085 | transmembrane transport | - | IEA | Process |
| GO:0071817 | MMXD complex | 20797633. | IDA | Component |
| GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 19154410. | IMP | Process |
| GO:1990830 | cellular response to leukemia inhibitory factor | - | IEA | Process |