
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| READ | |||
| STAD |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| SARC | ||
| HNSC | ||
| SKCM | ||
| BRCA | ||
| KIRP | ||
| PAAD | ||
| BLCA | ||
| CESC | ||
| UCEC | ||
| LGG | ||
| THCA | ||
| LUAD | ||
| OV |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000262630 | zf-C2H2 | PF00096.26 | 3.2e-08 | 1 | 1 |
| ENSP00000376035 | zf-C2H2 | PF00096.26 | 3.2e-08 | 1 | 1 |
| ENSP00000262630 | zf-met | PF12874.7 | 0.00058 | 1 | 1 |
| ENSP00000376035 | zf-met | PF12874.7 | 0.00058 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000442282 | ZBTB32-204 | 691 | - | - | - (aa) | - | - |
| ENST00000426659 | ZBTB32-203 | 469 | - | ENSP00000466524 | 71 (aa) | - | K7EMJ1 |
| ENST00000451726 | ZBTB32-205 | 355 | - | - | - (aa) | - | - |
| ENST00000392197 | ZBTB32-202 | 2068 | XM_017026591 | ENSP00000376035 | 487 (aa) | XP_016882080 | Q9Y2Y4 |
| ENST00000262630 | ZBTB32-201 | 1960 | XM_011526718 | ENSP00000262630 | 487 (aa) | XP_011525020 | Q9Y2Y4 |
| ENST00000481182 | ZBTB32-206 | 1733 | - | ENSP00000433657 | 302 (aa) | - | A0A0C4DGF1 |
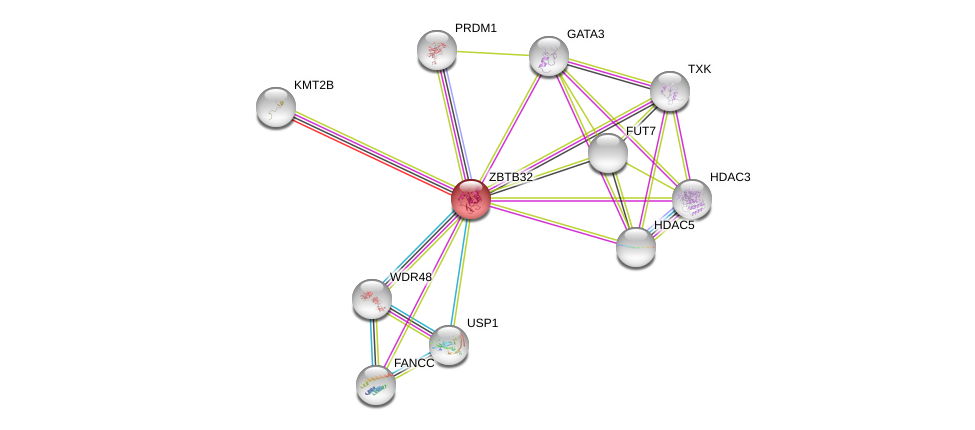
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000011590 | ZBTB32 | 99 | 79.261 | ENSAMEG00000000589 | ZBTB32 | 95 | 79.399 | Ailuropoda_melanoleuca |
| ENSG00000011590 | ZBTB32 | 100 | 94.366 | ENSANAG00000034122 | ZBTB32 | 96 | 89.079 | Aotus_nancymaae |
| ENSG00000011590 | ZBTB32 | 99 | 78.571 | ENSBTAG00000023611 | ZBTB32 | 96 | 76.119 | Bos_taurus |
| ENSG00000011590 | ZBTB32 | 100 | 88.732 | ENSCJAG00000046270 | ZBTB32 | 96 | 87.152 | Callithrix_jacchus |
| ENSG00000011590 | ZBTB32 | 100 | 81.690 | ENSCAFG00020018613 | ZBTB32 | 96 | 78.205 | Canis_lupus_dingo |
| ENSG00000011590 | ZBTB32 | 99 | 77.143 | ENSCHIG00000004618 | ZBTB32 | 96 | 76.546 | Capra_hircus |
| ENSG00000011590 | ZBTB32 | 99 | 82.857 | ENSTSYG00000008895 | ZBTB32 | 96 | 78.465 | Carlito_syrichta |
| ENSG00000011590 | ZBTB32 | 100 | 94.366 | ENSCCAG00000000774 | ZBTB32 | 96 | 88.437 | Cebus_capucinus |
| ENSG00000011590 | ZBTB32 | 100 | 98.592 | ENSCATG00000019611 | ZBTB32 | 96 | 95.075 | Cercocebus_atys |
| ENSG00000011590 | ZBTB32 | 100 | 98.592 | ENSCSAG00000004395 | ZBTB32 | 100 | 96.304 | Chlorocebus_sabaeus |
| ENSG00000011590 | ZBTB32 | 100 | 74.648 | ENSCHOG00000001017 | - | 60 | 84.828 | Choloepus_hoffmanni |
| ENSG00000011590 | ZBTB32 | 100 | 95.775 | ENSCANG00000031543 | ZBTB32 | 96 | 94.861 | Colobus_angolensis_palliatus |
| ENSG00000011590 | ZBTB32 | 99 | 66.736 | ENSCGRG00001018211 | Zbtb32 | 94 | 67.097 | Cricetulus_griseus_chok1gshd |
| ENSG00000011590 | ZBTB32 | 100 | 78.615 | ENSEASG00005001159 | ZBTB32 | 96 | 78.678 | Equus_asinus_asinus |
| ENSG00000011590 | ZBTB32 | 100 | 78.411 | ENSECAG00000005671 | ZBTB32 | 96 | 78.465 | Equus_caballus |
| ENSG00000011590 | ZBTB32 | 100 | 74.438 | ENSFCAG00000006380 | ZBTB32 | 96 | 74.359 | Felis_catus |
| ENSG00000011590 | ZBTB32 | 99 | 96.990 | ENSGGOG00000016789 | ZBTB32 | 96 | 94.658 | Gorilla_gorilla |
| ENSG00000011590 | ZBTB32 | 100 | 77.465 | ENSLAFG00000018739 | ZBTB32 | 96 | 73.013 | Loxodonta_africana |
| ENSG00000011590 | ZBTB32 | 100 | 98.592 | ENSMFAG00000034114 | ZBTB32 | 96 | 95.075 | Macaca_fascicularis |
| ENSG00000011590 | ZBTB32 | 100 | 97.183 | ENSMMUG00000002375 | ZBTB32 | 96 | 95.075 | Macaca_mulatta |
| ENSG00000011590 | ZBTB32 | 100 | 98.592 | ENSMNEG00000036607 | ZBTB32 | 96 | 95.289 | Macaca_nemestrina |
| ENSG00000011590 | ZBTB32 | 100 | 97.183 | ENSMLEG00000024571 | ZBTB32 | 96 | 94.004 | Mandrillus_leucophaeus |
| ENSG00000011590 | ZBTB32 | 99 | 88.571 | ENSMICG00000045992 | ZBTB32 | 96 | 81.702 | Microcebus_murinus |
| ENSG00000011590 | ZBTB32 | 100 | 76.056 | ENSMOCG00000004626 | Zbtb32 | 95 | 67.591 | Microtus_ochrogaster |
| ENSG00000011590 | ZBTB32 | 100 | 70.423 | MGP_CAROLIEiJ_G0029532 | Zbtb32 | 79 | 96.250 | Mus_caroli |
| ENSG00000011590 | ZBTB32 | 100 | 73.239 | ENSMUSG00000006310 | Zbtb32 | 79 | 96.250 | Mus_musculus |
| ENSG00000011590 | ZBTB32 | 100 | 73.239 | MGP_SPRETEiJ_G0030628 | Zbtb32 | 79 | 96.250 | Mus_spretus |
| ENSG00000011590 | ZBTB32 | 100 | 78.873 | ENSMPUG00000006234 | ZBTB32 | 95 | 72.318 | Mustela_putorius_furo |
| ENSG00000011590 | ZBTB32 | 100 | 84.507 | ENSMLUG00000011396 | ZBTB32 | 95 | 76.170 | Myotis_lucifugus |
| ENSG00000011590 | ZBTB32 | 100 | 97.741 | ENSNLEG00000011302 | ZBTB32 | 100 | 97.741 | Nomascus_leucogenys |
| ENSG00000011590 | ZBTB32 | 97 | 69.565 | ENSOPRG00000001316 | ZBTB32 | 96 | 59.873 | Ochotona_princeps |
| ENSG00000011590 | ZBTB32 | 100 | 84.507 | ENSOGAG00000004155 | ZBTB32 | 96 | 80.638 | Otolemur_garnettii |
| ENSG00000011590 | ZBTB32 | 100 | 100.000 | ENSPPAG00000031211 | ZBTB32 | 100 | 98.973 | Pan_paniscus |
| ENSG00000011590 | ZBTB32 | 100 | 74.438 | ENSPPRG00000008152 | ZBTB32 | 96 | 74.359 | Panthera_pardus |
| ENSG00000011590 | ZBTB32 | 100 | 74.642 | ENSPTIG00000021084 | ZBTB32 | 96 | 74.573 | Panthera_tigris_altaica |
| ENSG00000011590 | ZBTB32 | 100 | 100.000 | ENSPTRG00000010857 | ZBTB32 | 100 | 98.563 | Pan_troglodytes |
| ENSG00000011590 | ZBTB32 | 100 | 98.592 | ENSPANG00000009043 | ZBTB32 | 96 | 95.717 | Papio_anubis |
| ENSG00000011590 | ZBTB32 | 100 | 74.648 | ENSPEMG00000018640 | Zbtb32 | 94 | 70.108 | Peromyscus_maniculatus_bairdii |
| ENSG00000011590 | ZBTB32 | 100 | 98.592 | ENSPPYG00000009876 | ZBTB32 | 100 | 97.536 | Pongo_abelii |
| ENSG00000011590 | ZBTB32 | 100 | 85.915 | ENSPCOG00000003160 | ZBTB32 | 96 | 82.128 | Propithecus_coquereli |
| ENSG00000011590 | ZBTB32 | 99 | 80.000 | ENSPVAG00000012825 | ZBTB32 | 100 | 76.638 | Pteropus_vampyrus |
| ENSG00000011590 | ZBTB32 | 99 | 75.714 | ENSRNOG00000042465 | Zbtb32 | 95 | 69.807 | Rattus_norvegicus |
| ENSG00000011590 | ZBTB32 | 100 | 97.183 | ENSRBIG00000037073 | ZBTB32 | 96 | 95.075 | Rhinopithecus_bieti |
| ENSG00000011590 | ZBTB32 | 100 | 94.366 | ENSSBOG00000025572 | - | 96 | 88.651 | Saimiri_boliviensis_boliviensis |
| ENSG00000011590 | ZBTB32 | 100 | 91.549 | ENSSBOG00000034407 | - | 96 | 72.163 | Saimiri_boliviensis_boliviensis |
| ENSG00000011590 | ZBTB32 | 100 | 80.282 | ENSSSCG00000002908 | ZBTB32 | 97 | 76.333 | Sus_scrofa |
| ENSG00000011590 | ZBTB32 | 100 | 76.065 | ENSTBEG00000006535 | ZBTB32 | 95 | 76.546 | Tupaia_belangeri |
| ENSG00000011590 | ZBTB32 | 100 | 80.282 | ENSTTRG00000002232 | ZBTB32 | 64 | 85.128 | Tursiops_truncatus |
| ENSG00000011590 | ZBTB32 | 99 | 79.055 | ENSUAMG00000020706 | ZBTB32 | 95 | 79.185 | Ursus_americanus |
| ENSG00000011590 | ZBTB32 | 100 | 81.690 | ENSUMAG00000020255 | - | 89 | 85.586 | Ursus_maritimus |
| ENSG00000011590 | ZBTB32 | 100 | 78.074 | ENSVVUG00000015089 | ZBTB32 | 93 | 78.205 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000122 | negative regulation of transcription by RNA polymerase II | 10572087. | TAS | Process |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | - | ISA | Function |
| GO:0003677 | DNA binding | - | ISS | Function |
| GO:0003714 | transcription corepressor activity | 10572087. | TAS | Function |
| GO:0005515 | protein binding | 23088713. | IPI | Function |
| GO:0005634 | nucleus | 10572087. | TAS | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0006366 | transcription by RNA polymerase II | 10572087. | TAS | Process |
| GO:0008270 | zinc ion binding | - | ISS | Function |