
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| KIRC | |||
| LGG | |||
| SARC | |||
| SKCM | |||
| TGCT | |||
| UVM |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| STAD | ||
| UCS | ||
| LUSC | ||
| ESCA | ||
| KIRP | ||
| COAD | ||
| LIHC | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000262577 | zf-CCCH | PF00642.24 | 3e-19 | 1 | 2 |
| ENSP00000262577 | zf-CCCH | PF00642.24 | 3e-19 | 2 | 2 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000528401 | ZC3H3-202 | 546 | - | ENSP00000436834 | 79 (aa) | - | H0YEY2 |
| ENST00000262577 | ZC3H3-201 | 3280 | XM_006716536 | ENSP00000262577 | 948 (aa) | XP_006716599 | Q8IXZ2 |
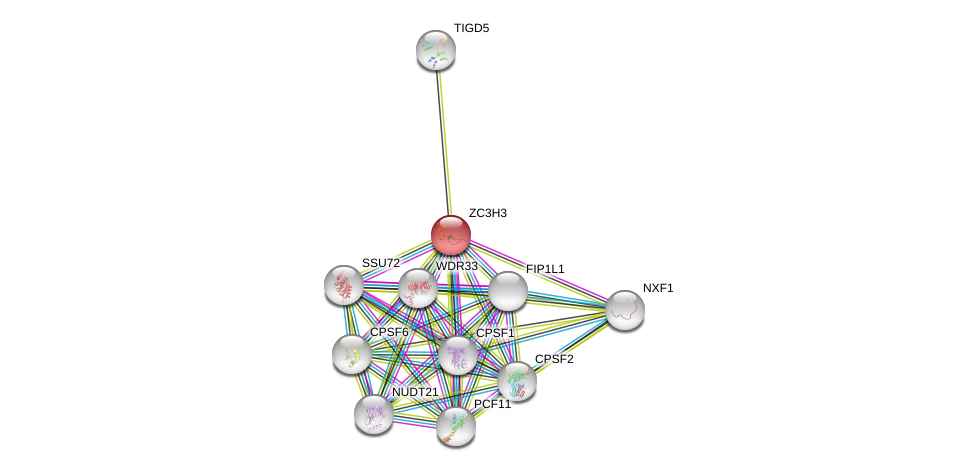
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000014164 | ZC3H3 | 62 | 43.023 | ENSAPOG00000011522 | zc3h3 | 64 | 42.776 | Acanthochromis_polyacanthus |
| ENSG00000014164 | ZC3H3 | 61 | 48.810 | ENSACIG00000005563 | zc3h3 | 65 | 48.276 | Amphilophus_citrinellus |
| ENSG00000014164 | ZC3H3 | 62 | 42.776 | ENSAOCG00000013763 | zc3h3 | 68 | 42.884 | Amphiprion_ocellaris |
| ENSG00000014164 | ZC3H3 | 60 | 42.942 | ENSAPEG00000020037 | zc3h3 | 60 | 43.056 | Amphiprion_percula |
| ENSG00000014164 | ZC3H3 | 86 | 34.399 | ENSATEG00000003718 | zc3h3 | 61 | 42.694 | Anabas_testudineus |
| ENSG00000014164 | ZC3H3 | 100 | 87.029 | ENSANAG00000018742 | ZC3H3 | 100 | 87.657 | Aotus_nancymaae |
| ENSG00000014164 | ZC3H3 | 76 | 45.238 | ENSACLG00000023035 | zc3h3 | 73 | 44.828 | Astatotilapia_calliptera |
| ENSG00000014164 | ZC3H3 | 91 | 72.248 | ENSBTAG00000021472 | ZC3H3 | 99 | 72.821 | Bos_taurus |
| ENSG00000014164 | ZC3H3 | 100 | 86.540 | ENSCJAG00000021439 | ZC3H3 | 100 | 86.961 | Callithrix_jacchus |
| ENSG00000014164 | ZC3H3 | 87 | 74.732 | ENSCAFG00000001299 | ZC3H3 | 100 | 75.566 | Canis_familiaris |
| ENSG00000014164 | ZC3H3 | 99 | 74.244 | ENSCAFG00020018654 | ZC3H3 | 100 | 75.182 | Canis_lupus_dingo |
| ENSG00000014164 | ZC3H3 | 100 | 73.291 | ENSCHIG00000018027 | ZC3H3 | 100 | 73.424 | Capra_hircus |
| ENSG00000014164 | ZC3H3 | 100 | 78.265 | ENSTSYG00000034906 | ZC3H3 | 100 | 78.892 | Carlito_syrichta |
| ENSG00000014164 | ZC3H3 | 97 | 66.881 | ENSCAPG00000013918 | ZC3H3 | 100 | 68.636 | Cavia_aperea |
| ENSG00000014164 | ZC3H3 | 100 | 73.849 | ENSCPOG00000006248 | ZC3H3 | 99 | 73.849 | Cavia_porcellus |
| ENSG00000014164 | ZC3H3 | 100 | 89.053 | ENSCCAG00000035267 | ZC3H3 | 100 | 89.053 | Cebus_capucinus |
| ENSG00000014164 | ZC3H3 | 100 | 93.263 | ENSCATG00000020900 | ZC3H3 | 100 | 93.684 | Cercocebus_atys |
| ENSG00000014164 | ZC3H3 | 100 | 74.790 | ENSCLAG00000009999 | ZC3H3 | 99 | 74.105 | Chinchilla_lanigera |
| ENSG00000014164 | ZC3H3 | 100 | 92.842 | ENSCSAG00000009074 | ZC3H3 | 99 | 93.579 | Chlorocebus_sabaeus |
| ENSG00000014164 | ZC3H3 | 100 | 46.496 | ENSCPBG00000019050 | ZC3H3 | 90 | 48.741 | Chrysemys_picta_bellii |
| ENSG00000014164 | ZC3H3 | 100 | 90.890 | ENSCANG00000032420 | ZC3H3 | 100 | 91.623 | Colobus_angolensis_palliatus |
| ENSG00000014164 | ZC3H3 | 100 | 72.414 | ENSCGRG00001016268 | Zc3h3 | 100 | 72.100 | Cricetulus_griseus_chok1gshd |
| ENSG00000014164 | ZC3H3 | 100 | 72.414 | ENSCGRG00000012889 | Zc3h3 | 100 | 72.100 | Cricetulus_griseus_crigri |
| ENSG00000014164 | ZC3H3 | 66 | 40.185 | ENSCSEG00000002583 | zc3h3 | 74 | 41.412 | Cynoglossus_semilaevis |
| ENSG00000014164 | ZC3H3 | 67 | 38.988 | ENSDARG00000090751 | zc3h3 | 89 | 35.821 | Danio_rerio |
| ENSG00000014164 | ZC3H3 | 88 | 67.947 | ENSDNOG00000031054 | ZC3H3 | 96 | 69.876 | Dasypus_novemcinctus |
| ENSG00000014164 | ZC3H3 | 98 | 71.030 | ENSDORG00000023832 | Zc3h3 | 100 | 71.030 | Dipodomys_ordii |
| ENSG00000014164 | ZC3H3 | 71 | 60.044 | ENSETEG00000014836 | - | 70 | 59.825 | Echinops_telfairi |
| ENSG00000014164 | ZC3H3 | 100 | 74.897 | ENSEASG00005009621 | ZC3H3 | 99 | 74.793 | Equus_asinus_asinus |
| ENSG00000014164 | ZC3H3 | 81 | 78.137 | ENSECAG00000012556 | ZC3H3 | 86 | 78.137 | Equus_caballus |
| ENSG00000014164 | ZC3H3 | 92 | 66.579 | ENSEEUG00000007986 | ZC3H3 | 91 | 66.579 | Erinaceus_europaeus |
| ENSG00000014164 | ZC3H3 | 64 | 42.163 | ENSELUG00000024269 | zc3h3 | 58 | 44.424 | Esox_lucius |
| ENSG00000014164 | ZC3H3 | 100 | 75.393 | ENSFCAG00000013238 | ZC3H3 | 100 | 76.021 | Felis_catus |
| ENSG00000014164 | ZC3H3 | 54 | 56.929 | ENSFALG00000009538 | ZC3H3 | 86 | 45.828 | Ficedula_albicollis |
| ENSG00000014164 | ZC3H3 | 100 | 72.670 | ENSFDAG00000017737 | ZC3H3 | 99 | 72.775 | Fukomys_damarensis |
| ENSG00000014164 | ZC3H3 | 61 | 44.141 | ENSFHEG00000017714 | zc3h3 | 61 | 46.512 | Fundulus_heteroclitus |
| ENSG00000014164 | ZC3H3 | 62 | 46.067 | ENSGAFG00000014360 | zc3h3 | 64 | 46.067 | Gambusia_affinis |
| ENSG00000014164 | ZC3H3 | 99 | 96.713 | ENSGGOG00000002833 | ZC3H3 | 100 | 97.425 | Gorilla_gorilla |
| ENSG00000014164 | ZC3H3 | 76 | 45.238 | ENSHBUG00000004627 | zc3h3 | 65 | 44.828 | Haplochromis_burtoni |
| ENSG00000014164 | ZC3H3 | 100 | 74.635 | ENSHGLG00000011234 | ZC3H3 | 99 | 74.635 | Heterocephalus_glaber_female |
| ENSG00000014164 | ZC3H3 | 92 | 72.027 | ENSHGLG00100006782 | ZC3H3 | 97 | 72.064 | Heterocephalus_glaber_male |
| ENSG00000014164 | ZC3H3 | 55 | 41.348 | ENSHCOG00000006579 | zc3h3 | 60 | 41.348 | Hippocampus_comes |
| ENSG00000014164 | ZC3H3 | 61 | 44.444 | ENSIPUG00000006438 | zc3h3 | 70 | 42.211 | Ictalurus_punctatus |
| ENSG00000014164 | ZC3H3 | 100 | 76.594 | ENSSTOG00000004358 | ZC3H3 | 99 | 76.071 | Ictidomys_tridecemlineatus |
| ENSG00000014164 | ZC3H3 | 100 | 70.231 | ENSJJAG00000023757 | Zc3h3 | 100 | 70.126 | Jaculus_jaculus |
| ENSG00000014164 | ZC3H3 | 64 | 45.455 | ENSKMAG00000017876 | zc3h3 | 64 | 45.455 | Kryptolebias_marmoratus |
| ENSG00000014164 | ZC3H3 | 77 | 42.090 | ENSLOCG00000008621 | ZC3H3 | 59 | 46.578 | Lepisosteus_oculatus |
| ENSG00000014164 | ZC3H3 | 100 | 93.263 | ENSMFAG00000036060 | ZC3H3 | 100 | 94.000 | Macaca_fascicularis |
| ENSG00000014164 | ZC3H3 | 100 | 93.263 | ENSMNEG00000029433 | ZC3H3 | 99 | 94.000 | Macaca_nemestrina |
| ENSG00000014164 | ZC3H3 | 100 | 93.158 | ENSMLEG00000038139 | ZC3H3 | 100 | 93.895 | Mandrillus_leucophaeus |
| ENSG00000014164 | ZC3H3 | 61 | 43.822 | ENSMAMG00000014787 | zc3h3 | 60 | 43.822 | Mastacembelus_armatus |
| ENSG00000014164 | ZC3H3 | 76 | 45.238 | ENSMZEG00005017290 | zc3h3 | 73 | 44.828 | Maylandia_zebra |
| ENSG00000014164 | ZC3H3 | 100 | 72.908 | ENSMAUG00000011009 | Zc3h3 | 100 | 72.594 | Mesocricetus_auratus |
| ENSG00000014164 | ZC3H3 | 100 | 79.768 | ENSMICG00000001100 | ZC3H3 | 100 | 79.768 | Microcebus_murinus |
| ENSG00000014164 | ZC3H3 | 100 | 71.369 | ENSMOCG00000000733 | Zc3h3 | 100 | 70.607 | Microtus_ochrogaster |
| ENSG00000014164 | ZC3H3 | 64 | 41.379 | ENSMMOG00000016823 | zc3h3 | 66 | 41.379 | Mola_mola |
| ENSG00000014164 | ZC3H3 | 88 | 61.483 | ENSMODG00000011385 | ZC3H3 | 94 | 61.493 | Monodelphis_domestica |
| ENSG00000014164 | ZC3H3 | 62 | 44.299 | ENSMALG00000014610 | zc3h3 | 61 | 44.299 | Monopterus_albus |
| ENSG00000014164 | ZC3H3 | 100 | 70.094 | MGP_CAROLIEiJ_G0019897 | Zc3h3 | 100 | 70.094 | Mus_caroli |
| ENSG00000014164 | ZC3H3 | 100 | 70.471 | ENSMUSG00000075600 | Zc3h3 | 100 | 70.471 | Mus_musculus |
| ENSG00000014164 | ZC3H3 | 100 | 69.688 | MGP_PahariEiJ_G0019905 | Zc3h3 | 100 | 70.417 | Mus_pahari |
| ENSG00000014164 | ZC3H3 | 100 | 70.471 | MGP_SPRETEiJ_G0020797 | Zc3h3 | 100 | 70.471 | Mus_spretus |
| ENSG00000014164 | ZC3H3 | 91 | 75.087 | ENSMPUG00000008357 | ZC3H3 | 97 | 75.087 | Mustela_putorius_furo |
| ENSG00000014164 | ZC3H3 | 100 | 73.563 | ENSNGAG00000008574 | Zc3h3 | 100 | 72.936 | Nannospalax_galili |
| ENSG00000014164 | ZC3H3 | 62 | 44.828 | ENSNBRG00000005454 | zc3h3 | 63 | 44.828 | Neolamprologus_brichardi |
| ENSG00000014164 | ZC3H3 | 100 | 91.588 | ENSNLEG00000001681 | ZC3H3 | 100 | 91.798 | Nomascus_leucogenys |
| ENSG00000014164 | ZC3H3 | 86 | 60.195 | ENSMEUG00000008554 | ZC3H3 | 100 | 56.854 | Notamacropus_eugenii |
| ENSG00000014164 | ZC3H3 | 100 | 72.228 | ENSODEG00000014239 | ZC3H3 | 100 | 72.936 | Octodon_degus |
| ENSG00000014164 | ZC3H3 | 96 | 50.429 | ENSOANG00000003534 | - | 98 | 50.701 | Ornithorhynchus_anatinus |
| ENSG00000014164 | ZC3H3 | 54 | 43.750 | ENSORLG00000026930 | zc3h3 | 58 | 43.038 | Oryzias_latipes |
| ENSG00000014164 | ZC3H3 | 62 | 42.884 | ENSORLG00020002734 | zc3h3 | 69 | 42.884 | Oryzias_latipes_hni |
| ENSG00000014164 | ZC3H3 | 61 | 43.307 | ENSORLG00015001257 | zc3h3 | 69 | 43.307 | Oryzias_latipes_hsok |
| ENSG00000014164 | ZC3H3 | 100 | 78.997 | ENSOGAG00000014907 | ZC3H3 | 100 | 78.668 | Otolemur_garnettii |
| ENSG00000014164 | ZC3H3 | 100 | 70.881 | ENSOARG00000000808 | ZC3H3 | 100 | 71.399 | Ovis_aries |
| ENSG00000014164 | ZC3H3 | 100 | 97.584 | ENSPPAG00000035455 | ZC3H3 | 87 | 97.584 | Pan_paniscus |
| ENSG00000014164 | ZC3H3 | 100 | 75.393 | ENSPPRG00000011707 | ZC3H3 | 100 | 76.021 | Panthera_pardus |
| ENSG00000014164 | ZC3H3 | 100 | 97.584 | ENSPTRG00000020655 | ZC3H3 | 100 | 97.584 | Pan_troglodytes |
| ENSG00000014164 | ZC3H3 | 100 | 92.632 | ENSPANG00000016578 | ZC3H3 | 100 | 93.368 | Papio_anubis |
| ENSG00000014164 | ZC3H3 | 100 | 73.117 | ENSPEMG00000013893 | Zc3h3 | 100 | 72.594 | Peromyscus_maniculatus_bairdii |
| ENSG00000014164 | ZC3H3 | 62 | 44.423 | ENSPFOG00000023405 | zc3h3 | 63 | 44.231 | Poecilia_formosa |
| ENSG00000014164 | ZC3H3 | 61 | 44.246 | ENSPLAG00000001686 | zc3h3 | 62 | 44.246 | Poecilia_latipinna |
| ENSG00000014164 | ZC3H3 | 77 | 42.697 | ENSPMEG00000002341 | zc3h3 | 63 | 44.423 | Poecilia_mexicana |
| ENSG00000014164 | ZC3H3 | 77 | 46.067 | ENSPREG00000008923 | zc3h3 | 72 | 46.067 | Poecilia_reticulata |
| ENSG00000014164 | ZC3H3 | 100 | 94.415 | ENSPPYG00000018935 | ZC3H3 | 99 | 94.942 | Pongo_abelii |
| ENSG00000014164 | ZC3H3 | 100 | 80.294 | ENSPCOG00000028174 | ZC3H3 | 100 | 81.027 | Propithecus_coquereli |
| ENSG00000014164 | ZC3H3 | 88 | 73.750 | ENSPVAG00000005979 | ZC3H3 | 88 | 74.375 | Pteropus_vampyrus |
| ENSG00000014164 | ZC3H3 | 76 | 45.238 | ENSPNYG00000023105 | zc3h3 | 65 | 44.828 | Pundamilia_nyererei |
| ENSG00000014164 | ZC3H3 | 100 | 92.955 | ENSRBIG00000039855 | ZC3H3 | 100 | 93.691 | Rhinopithecus_bieti |
| ENSG00000014164 | ZC3H3 | 100 | 92.955 | ENSRROG00000040493 | ZC3H3 | 100 | 93.691 | Rhinopithecus_roxellana |
| ENSG00000014164 | ZC3H3 | 100 | 87.945 | ENSSBOG00000021819 | ZC3H3 | 100 | 88.470 | Saimiri_boliviensis_boliviensis |
| ENSG00000014164 | ZC3H3 | 62 | 46.429 | ENSSDUG00000021871 | zc3h3 | 62 | 46.429 | Seriola_dumerili |
| ENSG00000014164 | ZC3H3 | 62 | 46.429 | ENSSLDG00000016166 | zc3h3 | 62 | 46.429 | Seriola_lalandi_dorsalis |
| ENSG00000014164 | ZC3H3 | 61 | 42.557 | ENSSPAG00000008518 | zc3h3 | 62 | 42.557 | Stegastes_partitus |
| ENSG00000014164 | ZC3H3 | 100 | 70.934 | ENSSSCG00000006956 | ZC3H3 | 99 | 71.653 | Sus_scrofa |
| ENSG00000014164 | ZC3H3 | 77 | 64.423 | ENSTBEG00000009096 | - | 85 | 64.423 | Tupaia_belangeri |
| ENSG00000014164 | ZC3H3 | 91 | 78.911 | ENSUAMG00000012919 | ZC3H3 | 97 | 79.606 | Ursus_americanus |
| ENSG00000014164 | ZC3H3 | 99 | 74.332 | ENSVVUG00000022609 | ZC3H3 | 98 | 75.586 | Vulpes_vulpes |
| ENSG00000014164 | ZC3H3 | 54 | 51.679 | ENSXETG00000004937 | zc3h3 | 91 | 41.812 | Xenopus_tropicalis |
| ENSG00000014164 | ZC3H3 | 53 | 42.777 | ENSXCOG00000005025 | zc3h3 | 66 | 42.777 | Xiphophorus_couchianus |
| ENSG00000014164 | ZC3H3 | 62 | 48.315 | ENSXMAG00000006422 | zc3h3 | 64 | 48.315 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 19274049. | ISM | Function |
| GO:0003674 | molecular_function | - | ND | Function |
| GO:0003677 | DNA binding | - | IEA | Function |
| GO:0005634 | nucleus | 16115198.19364924. | IDA | Component |
| GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex | - | IEA | Component |
| GO:0006357 | regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0006378 | mRNA polyadenylation | 19364924. | IMP | Process |
| GO:0010793 | regulation of mRNA export from nucleus | 19364924. | IMP | Process |
| GO:0016973 | poly(A)+ mRNA export from nucleus | 21873635. | IBA | Process |
| GO:0016973 | poly(A)+ mRNA export from nucleus | 19364924. | IMP | Process |
| GO:0032927 | positive regulation of activin receptor signaling pathway | - | IEA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0070412 | R-SMAD binding | - | IEA | Function |