
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| KIRP | |||
| LUAD | |||
| LUSC | |||
| OV | |||
| PAAD | |||
| READ | |||
| TGCT | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| LUSC | ||
| ESCA | ||
| PAAD | ||
| BLCA | ||
| CESC | ||
| GBM | ||
| LIHC | ||
| THCA | ||
| UVM | ||
| OV |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000263277 | MMR_HSR1 | PF01926.23 | 7.2e-07 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000596225 | EHD2-204 | 538 | - | ENSP00000471766 | 30 (aa) | - | M0R1B9 |
| ENST00000540884 | EHD2-203 | 1215 | - | - | - (aa) | - | - |
| ENST00000538399 | EHD2-202 | 1766 | - | ENSP00000439036 | 407 (aa) | - | Q9NZN4 |
| ENST00000263277 | EHD2-201 | 3585 | - | ENSP00000263277 | 543 (aa) | - | Q9NZN4 |
| ENST00000602215 | EHD2-205 | 234 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04144 | Endocytosis | KEGG |
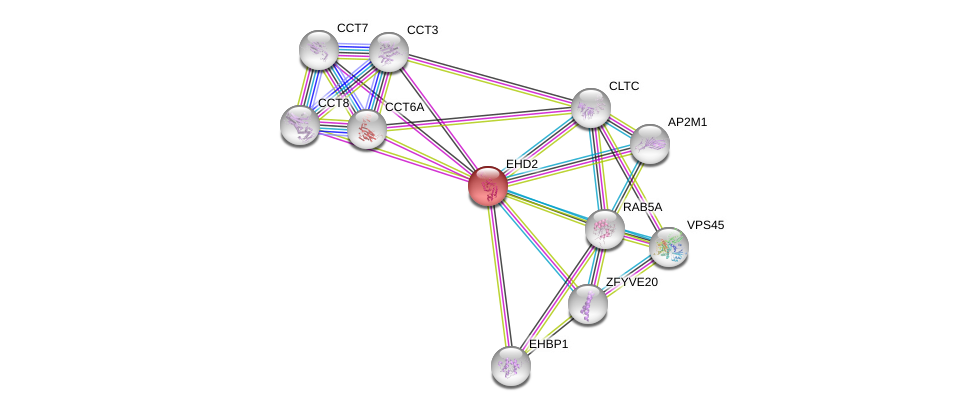
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000024422 | EHD2 | 99 | 81.728 | ENSAPOG00000012705 | ehd2b | 98 | 80.755 | Acanthochromis_polyacanthus |
| ENSG00000024422 | EHD2 | 99 | 65.764 | ENSAPOG00000021608 | - | 97 | 66.730 | Acanthochromis_polyacanthus |
| ENSG00000024422 | EHD2 | 100 | 99.079 | ENSAMEG00000015263 | EHD2 | 100 | 99.079 | Ailuropoda_melanoleuca |
| ENSG00000024422 | EHD2 | 99 | 65.926 | ENSACIG00000011074 | - | 99 | 65.000 | Amphilophus_citrinellus |
| ENSG00000024422 | EHD2 | 99 | 81.975 | ENSAOCG00000006422 | EHD2 | 98 | 81.132 | Amphiprion_ocellaris |
| ENSG00000024422 | EHD2 | 99 | 65.517 | ENSAOCG00000002163 | - | 97 | 66.540 | Amphiprion_ocellaris |
| ENSG00000024422 | EHD2 | 99 | 81.975 | ENSAPEG00000013956 | ehd2b | 98 | 81.132 | Amphiprion_percula |
| ENSG00000024422 | EHD2 | 99 | 65.025 | ENSAPEG00000019382 | - | 97 | 66.160 | Amphiprion_percula |
| ENSG00000024422 | EHD2 | 98 | 65.337 | ENSATEG00000004166 | - | 96 | 66.411 | Anabas_testudineus |
| ENSG00000024422 | EHD2 | 100 | 99.754 | ENSANAG00000031077 | EHD2 | 100 | 99.632 | Aotus_nancymaae |
| ENSG00000024422 | EHD2 | 99 | 80.494 | ENSACLG00000021088 | ehd2b | 99 | 79.511 | Astatotilapia_calliptera |
| ENSG00000024422 | EHD2 | 99 | 64.444 | ENSACLG00000005241 | - | 97 | 65.143 | Astatotilapia_calliptera |
| ENSG00000024422 | EHD2 | 99 | 71.776 | ENSAMXG00000013748 | ehd2a | 95 | 72.316 | Astyanax_mexicanus |
| ENSG00000024422 | EHD2 | 100 | 98.771 | ENSBTAG00000021191 | EHD2 | 100 | 98.711 | Bos_taurus |
| ENSG00000024422 | EHD2 | 100 | 99.754 | ENSCJAG00000011415 | EHD2 | 100 | 99.632 | Callithrix_jacchus |
| ENSG00000024422 | EHD2 | 100 | 99.263 | ENSCAFG00000004095 | EHD2 | 100 | 98.899 | Canis_familiaris |
| ENSG00000024422 | EHD2 | 100 | 99.263 | ENSCAFG00020000226 | EHD2 | 100 | 99.263 | Canis_lupus_dingo |
| ENSG00000024422 | EHD2 | 100 | 98.527 | ENSCHIG00000011896 | EHD2 | 100 | 98.527 | Capra_hircus |
| ENSG00000024422 | EHD2 | 100 | 97.422 | ENSCAPG00000007033 | EHD2 | 100 | 97.422 | Cavia_aperea |
| ENSG00000024422 | EHD2 | 100 | 97.422 | ENSCPOG00000026417 | EHD2 | 100 | 97.422 | Cavia_porcellus |
| ENSG00000024422 | EHD2 | 100 | 99.754 | ENSCCAG00000035777 | EHD2 | 100 | 99.632 | Cebus_capucinus |
| ENSG00000024422 | EHD2 | 100 | 97.974 | ENSCLAG00000001756 | EHD2 | 100 | 97.974 | Chinchilla_lanigera |
| ENSG00000024422 | EHD2 | 100 | 99.816 | ENSCSAG00000002930 | EHD2 | 100 | 99.816 | Chlorocebus_sabaeus |
| ENSG00000024422 | EHD2 | 100 | 85.012 | ENSCPBG00000009041 | EHD2 | 100 | 82.320 | Chrysemys_picta_bellii |
| ENSG00000024422 | EHD2 | 100 | 99.816 | ENSCANG00000040566 | EHD2 | 100 | 99.816 | Colobus_angolensis_palliatus |
| ENSG00000024422 | EHD2 | 100 | 98.034 | ENSCGRG00001018635 | Ehd2 | 100 | 97.790 | Cricetulus_griseus_chok1gshd |
| ENSG00000024422 | EHD2 | 99 | 80.049 | ENSCSEG00000007149 | ehd2b | 98 | 78.908 | Cynoglossus_semilaevis |
| ENSG00000024422 | EHD2 | 99 | 64.390 | ENSCSEG00000001675 | - | 97 | 65.849 | Cynoglossus_semilaevis |
| ENSG00000024422 | EHD2 | 99 | 82.222 | ENSCVAG00000002381 | ehd2b | 98 | 81.132 | Cyprinodon_variegatus |
| ENSG00000024422 | EHD2 | 99 | 65.271 | ENSCVAG00000018998 | - | 97 | 65.970 | Cyprinodon_variegatus |
| ENSG00000024422 | EHD2 | 99 | 83.047 | ENSDARG00000040362 | ehd2b | 98 | 81.955 | Danio_rerio |
| ENSG00000024422 | EHD2 | 99 | 68.627 | ENSDARG00000035137 | ehd2a | 97 | 69.288 | Danio_rerio |
| ENSG00000024422 | EHD2 | 100 | 98.527 | ENSDORG00000011700 | Ehd2 | 100 | 98.527 | Dipodomys_ordii |
| ENSG00000024422 | EHD2 | 100 | 98.895 | ENSECAG00000028265 | EHD2 | 100 | 98.895 | Equus_caballus |
| ENSG00000024422 | EHD2 | 56 | 96.393 | ENSEEUG00000000956 | EHD2 | 100 | 96.393 | Erinaceus_europaeus |
| ENSG00000024422 | EHD2 | 99 | 67.079 | ENSELUG00000019299 | ehd2a | 97 | 67.557 | Esox_lucius |
| ENSG00000024422 | EHD2 | 99 | 80.835 | ENSELUG00000021876 | ehd2b | 98 | 80.451 | Esox_lucius |
| ENSG00000024422 | EHD2 | 100 | 98.527 | ENSFCAG00000029117 | EHD2 | 100 | 98.527 | Felis_catus |
| ENSG00000024422 | EHD2 | 100 | 97.543 | ENSFDAG00000015675 | EHD2 | 100 | 97.552 | Fukomys_damarensis |
| ENSG00000024422 | EHD2 | 99 | 81.481 | ENSFHEG00000020637 | ehd2b | 98 | 80.943 | Fundulus_heteroclitus |
| ENSG00000024422 | EHD2 | 99 | 66.010 | ENSFHEG00000002126 | - | 97 | 66.540 | Fundulus_heteroclitus |
| ENSG00000024422 | EHD2 | 98 | 64.356 | ENSGMOG00000000211 | - | 97 | 65.217 | Gadus_morhua |
| ENSG00000024422 | EHD2 | 99 | 79.218 | ENSGMOG00000013367 | ehd2b | 98 | 79.401 | Gadus_morhua |
| ENSG00000024422 | EHD2 | 100 | 78.133 | ENSGALG00000036498 | EHD2 | 100 | 77.901 | Gallus_gallus |
| ENSG00000024422 | EHD2 | 99 | 64.778 | ENSGAFG00000004457 | ehd2a | 97 | 65.779 | Gambusia_affinis |
| ENSG00000024422 | EHD2 | 99 | 63.636 | ENSGACG00000007320 | - | 97 | 64.326 | Gasterosteus_aculeatus |
| ENSG00000024422 | EHD2 | 99 | 80.247 | ENSGACG00000013023 | ehd2b | 98 | 79.434 | Gasterosteus_aculeatus |
| ENSG00000024422 | EHD2 | 100 | 85.749 | ENSGAGG00000022066 | EHD2 | 100 | 82.873 | Gopherus_agassizii |
| ENSG00000024422 | EHD2 | 100 | 100.000 | ENSGGOG00000002346 | EHD2 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000024422 | EHD2 | 99 | 80.494 | ENSHBUG00000009900 | ehd2b | 98 | 79.434 | Haplochromis_burtoni |
| ENSG00000024422 | EHD2 | 99 | 64.691 | ENSHBUG00000008791 | - | 97 | 65.333 | Haplochromis_burtoni |
| ENSG00000024422 | EHD2 | 100 | 97.974 | ENSHGLG00000017825 | EHD2 | 100 | 97.974 | Heterocephalus_glaber_female |
| ENSG00000024422 | EHD2 | 100 | 97.974 | ENSHGLG00100017344 | EHD2 | 100 | 97.974 | Heterocephalus_glaber_male |
| ENSG00000024422 | EHD2 | 99 | 64.179 | ENSHCOG00000012768 | - | 98 | 64.179 | Hippocampus_comes |
| ENSG00000024422 | EHD2 | 99 | 80.835 | ENSHCOG00000017683 | ehd2b | 97 | 79.887 | Hippocampus_comes |
| ENSG00000024422 | EHD2 | 99 | 71.499 | ENSIPUG00000024057 | ehd2a | 98 | 70.588 | Ictalurus_punctatus |
| ENSG00000024422 | EHD2 | 99 | 81.818 | ENSIPUG00000000204 | Ehd2 | 98 | 81.391 | Ictalurus_punctatus |
| ENSG00000024422 | EHD2 | 100 | 98.526 | ENSJJAG00000008621 | Ehd2 | 100 | 97.790 | Jaculus_jaculus |
| ENSG00000024422 | EHD2 | 99 | 64.778 | ENSKMAG00000003332 | - | 97 | 65.209 | Kryptolebias_marmoratus |
| ENSG00000024422 | EHD2 | 99 | 81.728 | ENSKMAG00000012447 | ehd2b | 98 | 80.755 | Kryptolebias_marmoratus |
| ENSG00000024422 | EHD2 | 99 | 79.856 | ENSLBEG00000018343 | ehd2b | 98 | 79.336 | Labrus_bergylta |
| ENSG00000024422 | EHD2 | 99 | 65.602 | ENSLBEG00000004961 | - | 97 | 66.603 | Labrus_bergylta |
| ENSG00000024422 | EHD2 | 79 | 89.286 | ENSLACG00000018332 | ehd2a | 95 | 83.934 | Latimeria_chalumnae |
| ENSG00000024422 | EHD2 | 100 | 99.816 | ENSMMUG00000040121 | EHD2 | 100 | 99.816 | Macaca_mulatta |
| ENSG00000024422 | EHD2 | 99 | 65.764 | ENSMAMG00000004890 | - | 97 | 66.730 | Mastacembelus_armatus |
| ENSG00000024422 | EHD2 | 99 | 64.444 | ENSMZEG00005005247 | - | 97 | 65.143 | Maylandia_zebra |
| ENSG00000024422 | EHD2 | 99 | 80.494 | ENSMZEG00005024162 | ehd2b | 98 | 79.434 | Maylandia_zebra |
| ENSG00000024422 | EHD2 | 100 | 97.789 | ENSMAUG00000010317 | Ehd2 | 100 | 97.606 | Mesocricetus_auratus |
| ENSG00000024422 | EHD2 | 100 | 96.685 | ENSMICG00000005571 | EHD2 | 100 | 96.685 | Microcebus_murinus |
| ENSG00000024422 | EHD2 | 100 | 98.034 | ENSMOCG00000007010 | Ehd2 | 100 | 97.238 | Microtus_ochrogaster |
| ENSG00000024422 | EHD2 | 98 | 64.090 | ENSMMOG00000009001 | - | 97 | 64.981 | Mola_mola |
| ENSG00000024422 | EHD2 | 99 | 80.494 | ENSMALG00000008756 | ehd2b | 98 | 79.623 | Monopterus_albus |
| ENSG00000024422 | EHD2 | 100 | 97.052 | MGP_CAROLIEiJ_G0029219 | Ehd2 | 100 | 100.000 | Mus_caroli |
| ENSG00000024422 | EHD2 | 100 | 97.052 | ENSMUSG00000074364 | Ehd2 | 100 | 100.000 | Mus_musculus |
| ENSG00000024422 | EHD2 | 100 | 97.297 | MGP_PahariEiJ_G0021075 | Ehd2 | 100 | 100.000 | Mus_pahari |
| ENSG00000024422 | EHD2 | 100 | 97.297 | MGP_SPRETEiJ_G0030299 | Ehd2 | 100 | 100.000 | Mus_spretus |
| ENSG00000024422 | EHD2 | 100 | 99.263 | ENSMPUG00000004466 | EHD2 | 100 | 99.079 | Mustela_putorius_furo |
| ENSG00000024422 | EHD2 | 100 | 99.017 | ENSNGAG00000018699 | Ehd2 | 100 | 98.527 | Nannospalax_galili |
| ENSG00000024422 | EHD2 | 99 | 64.691 | ENSNBRG00000002925 | ehd2a | 99 | 63.904 | Neolamprologus_brichardi |
| ENSG00000024422 | EHD2 | 100 | 89.435 | ENSMEUG00000000445 | EHD2 | 100 | 90.110 | Notamacropus_eugenii |
| ENSG00000024422 | EHD2 | 100 | 97.790 | ENSODEG00000006641 | EHD2 | 100 | 97.790 | Octodon_degus |
| ENSG00000024422 | EHD2 | 99 | 64.373 | ENSONIG00000005597 | - | 99 | 64.088 | Oreochromis_niloticus |
| ENSG00000024422 | EHD2 | 99 | 80.741 | ENSONIG00000004051 | ehd2b | 98 | 79.623 | Oreochromis_niloticus |
| ENSG00000024422 | EHD2 | 99 | 81.481 | ENSORLG00000002244 | ehd2b | 98 | 80.189 | Oryzias_latipes |
| ENSG00000024422 | EHD2 | 98 | 64.339 | ENSORLG00000012013 | - | 96 | 64.627 | Oryzias_latipes |
| ENSG00000024422 | EHD2 | 98 | 64.339 | ENSORLG00020013250 | - | 96 | 64.627 | Oryzias_latipes_hni |
| ENSG00000024422 | EHD2 | 99 | 81.235 | ENSORLG00020017051 | ehd2b | 98 | 80.000 | Oryzias_latipes_hni |
| ENSG00000024422 | EHD2 | 98 | 64.589 | ENSORLG00015021510 | - | 96 | 64.818 | Oryzias_latipes_hsok |
| ENSG00000024422 | EHD2 | 99 | 81.481 | ENSORLG00015005729 | ehd2 | 98 | 80.189 | Oryzias_latipes_hsok |
| ENSG00000024422 | EHD2 | 98 | 64.589 | ENSOMEG00000003975 | - | 96 | 64.818 | Oryzias_melastigma |
| ENSG00000024422 | EHD2 | 99 | 81.481 | ENSOMEG00000001261 | ehd2b | 98 | 81.132 | Oryzias_melastigma |
| ENSG00000024422 | EHD2 | 100 | 98.895 | ENSOGAG00000033142 | EHD2 | 100 | 98.895 | Otolemur_garnettii |
| ENSG00000024422 | EHD2 | 97 | 97.148 | ENSOARG00000011391 | EHD2 | 89 | 97.148 | Ovis_aries |
| ENSG00000024422 | EHD2 | 100 | 96.317 | ENSPPAG00000039669 | EHD2 | 100 | 96.317 | Pan_paniscus |
| ENSG00000024422 | EHD2 | 100 | 98.771 | ENSPPRG00000008148 | EHD2 | 100 | 98.711 | Panthera_pardus |
| ENSG00000024422 | EHD2 | 100 | 98.771 | ENSPTIG00000018608 | EHD2 | 100 | 98.711 | Panthera_tigris_altaica |
| ENSG00000024422 | EHD2 | 100 | 99.632 | ENSPTRG00000011214 | EHD2 | 100 | 99.632 | Pan_troglodytes |
| ENSG00000024422 | EHD2 | 100 | 99.632 | ENSPTRG00000046930 | EHD2 | 100 | 99.632 | Pan_troglodytes |
| ENSG00000024422 | EHD2 | 100 | 99.816 | ENSPANG00000026345 | EHD2 | 100 | 99.816 | Papio_anubis |
| ENSG00000024422 | EHD2 | 99 | 82.222 | ENSPKIG00000013262 | EHD2 | 99 | 80.258 | Paramormyrops_kingsleyae |
| ENSG00000024422 | EHD2 | 99 | 78.818 | ENSPKIG00000016765 | ehd2a | 99 | 76.796 | Paramormyrops_kingsleyae |
| ENSG00000024422 | EHD2 | 99 | 64.951 | ENSPMGG00000013285 | - | 97 | 65.530 | Periophthalmus_magnuspinnatus |
| ENSG00000024422 | EHD2 | 99 | 80.741 | ENSPMGG00000015379 | ehd2b | 99 | 80.337 | Periophthalmus_magnuspinnatus |
| ENSG00000024422 | EHD2 | 100 | 94.103 | ENSPCIG00000016714 | EHD2 | 100 | 93.773 | Phascolarctos_cinereus |
| ENSG00000024422 | EHD2 | 99 | 82.222 | ENSPFOG00000011756 | ehd2b | 98 | 81.698 | Poecilia_formosa |
| ENSG00000024422 | EHD2 | 99 | 63.300 | ENSPFOG00000012774 | - | 96 | 64.639 | Poecilia_formosa |
| ENSG00000024422 | EHD2 | 99 | 63.547 | ENSPLAG00000000301 | - | 97 | 64.829 | Poecilia_latipinna |
| ENSG00000024422 | EHD2 | 99 | 82.469 | ENSPLAG00000020534 | ehd2b | 98 | 81.887 | Poecilia_latipinna |
| ENSG00000024422 | EHD2 | 99 | 63.793 | ENSPMEG00000024037 | - | 97 | 65.019 | Poecilia_mexicana |
| ENSG00000024422 | EHD2 | 99 | 82.222 | ENSPMEG00000008030 | ehd2b | 98 | 81.698 | Poecilia_mexicana |
| ENSG00000024422 | EHD2 | 99 | 63.300 | ENSPREG00000017608 | ehd2a | 97 | 64.639 | Poecilia_reticulata |
| ENSG00000024422 | EHD2 | 99 | 82.222 | ENSPREG00000016332 | ehd2b | 98 | 81.887 | Poecilia_reticulata |
| ENSG00000024422 | EHD2 | 100 | 99.816 | ENSPPYG00000010180 | EHD2 | 90 | 99.816 | Pongo_abelii |
| ENSG00000024422 | EHD2 | 56 | 98.033 | ENSPCAG00000007707 | EHD2 | 100 | 98.033 | Procavia_capensis |
| ENSG00000024422 | EHD2 | 100 | 98.711 | ENSPCOG00000010027 | EHD2 | 100 | 98.711 | Propithecus_coquereli |
| ENSG00000024422 | EHD2 | 100 | 93.923 | ENSPVAG00000014670 | EHD2 | 100 | 93.923 | Pteropus_vampyrus |
| ENSG00000024422 | EHD2 | 99 | 80.494 | ENSPNYG00000007864 | ehd2b | 98 | 79.434 | Pundamilia_nyererei |
| ENSG00000024422 | EHD2 | 99 | 64.444 | ENSPNYG00000009593 | - | 97 | 65.143 | Pundamilia_nyererei |
| ENSG00000024422 | EHD2 | 99 | 81.311 | ENSPNAG00000026764 | ehd2b | 98 | 80.669 | Pygocentrus_nattereri |
| ENSG00000024422 | EHD2 | 99 | 72.549 | ENSPNAG00000016978 | ehd2a | 97 | 71.719 | Pygocentrus_nattereri |
| ENSG00000024422 | EHD2 | 99 | 82.512 | ENSPNAG00000026560 | ehd2b | 98 | 81.579 | Pygocentrus_nattereri |
| ENSG00000024422 | EHD2 | 100 | 97.789 | ENSRNOG00000011346 | Ehd2 | 100 | 97.053 | Rattus_norvegicus |
| ENSG00000024422 | EHD2 | 100 | 99.816 | ENSRBIG00000035965 | EHD2 | 100 | 99.816 | Rhinopithecus_bieti |
| ENSG00000024422 | EHD2 | 100 | 99.816 | ENSRROG00000043148 | EHD2 | 100 | 99.816 | Rhinopithecus_roxellana |
| ENSG00000024422 | EHD2 | 100 | 99.263 | ENSSBOG00000025594 | EHD2 | 100 | 99.263 | Saimiri_boliviensis_boliviensis |
| ENSG00000024422 | EHD2 | 90 | 73.747 | ENSSFOG00015006753 | ehd2a | 95 | 81.638 | Scleropages_formosus |
| ENSG00000024422 | EHD2 | 99 | 82.020 | ENSSFOG00015023056 | ehd2 | 100 | 79.336 | Scleropages_formosus |
| ENSG00000024422 | EHD2 | 99 | 65.764 | ENSSMAG00000020341 | - | 97 | 67.110 | Scophthalmus_maximus |
| ENSG00000024422 | EHD2 | 99 | 80.000 | ENSSMAG00000020558 | ehd2b | 98 | 79.434 | Scophthalmus_maximus |
| ENSG00000024422 | EHD2 | 99 | 66.502 | ENSSDUG00000020042 | - | 97 | 67.823 | Seriola_dumerili |
| ENSG00000024422 | EHD2 | 99 | 81.481 | ENSSDUG00000000141 | ehd2b | 98 | 80.755 | Seriola_dumerili |
| ENSG00000024422 | EHD2 | 99 | 81.481 | ENSSLDG00000005368 | ehd2b | 98 | 80.755 | Seriola_lalandi_dorsalis |
| ENSG00000024422 | EHD2 | 99 | 66.995 | ENSSLDG00000008515 | - | 97 | 67.681 | Seriola_lalandi_dorsalis |
| ENSG00000024422 | EHD2 | 100 | 83.824 | ENSSPUG00000004401 | EHD2 | 100 | 81.768 | Sphenodon_punctatus |
| ENSG00000024422 | EHD2 | 99 | 65.271 | ENSSPAG00000006737 | - | 97 | 66.540 | Stegastes_partitus |
| ENSG00000024422 | EHD2 | 99 | 81.481 | ENSSPAG00000018164 | ehd2b | 98 | 80.566 | Stegastes_partitus |
| ENSG00000024422 | EHD2 | 100 | 99.263 | ENSSSCG00000023371 | EHD2 | 100 | 99.079 | Sus_scrofa |
| ENSG00000024422 | EHD2 | 99 | 80.693 | ENSTRUG00000010128 | ehd2b | 98 | 79.773 | Takifugu_rubripes |
| ENSG00000024422 | EHD2 | 99 | 81.235 | ENSTNIG00000016284 | ehd2b | 97 | 80.377 | Tetraodon_nigroviridis |
| ENSG00000024422 | EHD2 | 100 | 97.790 | ENSTTRG00000011850 | EHD2 | 100 | 97.790 | Tursiops_truncatus |
| ENSG00000024422 | EHD2 | 100 | 99.263 | ENSUAMG00000028253 | EHD2 | 100 | 99.263 | Ursus_americanus |
| ENSG00000024422 | EHD2 | 50 | 98.897 | ENSUMAG00000023829 | - | 74 | 98.897 | Ursus_maritimus |
| ENSG00000024422 | EHD2 | 100 | 99.263 | ENSVVUG00000023369 | EHD2 | 88 | 99.263 | Vulpes_vulpes |
| ENSG00000024422 | EHD2 | 100 | 82.801 | ENSXETG00000002783 | ehd2 | 100 | 80.295 | Xenopus_tropicalis |
| ENSG00000024422 | EHD2 | 99 | 64.532 | ENSXCOG00000012882 | - | 98 | 65.209 | Xiphophorus_couchianus |
| ENSG00000024422 | EHD2 | 99 | 82.469 | ENSXCOG00000008602 | ehd2b | 99 | 80.591 | Xiphophorus_couchianus |
| ENSG00000024422 | EHD2 | 99 | 64.373 | ENSXMAG00000010771 | ehd2a | 97 | 65.465 | Xiphophorus_maculatus |
| ENSG00000024422 | EHD2 | 99 | 82.222 | ENSXMAG00000029134 | ehd2b | 98 | 81.509 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003676 | nucleic acid binding | 10673336. | TAS | Function |
| GO:0005509 | calcium ion binding | - | IEA | Function |
| GO:0005515 | protein binding | 17233914.25588833. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005634 | nucleus | 10673336. | TAS | Component |
| GO:0005829 | cytosol | - | ISS | Component |
| GO:0005886 | plasma membrane | - | IDA | Component |
| GO:0005886 | plasma membrane | - | ISS | Component |
| GO:0005901 | caveola | 24013648. | IDA | Component |
| GO:0006897 | endocytosis | - | IEA | Process |
| GO:0007596 | blood coagulation | - | TAS | Process |
| GO:0010008 | endosome membrane | - | TAS | Component |
| GO:0015630 | microtubule cytoskeleton | - | IDA | Component |
| GO:0016787 | hydrolase activity | - | IEA | Function |
| GO:0019898 | extrinsic component of membrane | - | ISS | Component |
| GO:0019904 | protein domain specific binding | - | IEA | Function |
| GO:0030866 | cortical actin cytoskeleton organization | - | IEA | Process |
| GO:0032456 | endocytic recycling | 17233914. | IGI | Process |
| GO:0042802 | identical protein binding | - | IEA | Function |
| GO:0045171 | intercellular bridge | - | IDA | Component |
| GO:0048471 | perinuclear region of cytoplasm | - | IEA | Component |
| GO:0055038 | recycling endosome membrane | 17233914. | IDA | Component |
| GO:0070062 | extracellular exosome | 19056867. | HDA | Component |
| GO:0072659 | protein localization to plasma membrane | - | ISS | Process |
| GO:0097320 | plasma membrane tubulation | - | ISS | Process |
| GO:1901741 | positive regulation of myoblast fusion | - | ISS | Process |
| GO:2001137 | positive regulation of endocytic recycling | - | ISS | Process |