
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| LAML | |||
| LUAD | |||
| PRAD |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| MESO | ||
| HNSC | ||
| ESCA | ||
| COAD | ||
| BLCA | ||
| READ | ||
| LAML | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| THCA |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 7941739 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000372813 | TOMM34-201 | 2062 | XM_005260254 | ENSP00000361900 | 309 (aa) | XP_005260311 | Q15785 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000025772 | Macular Degeneration | 1.268967E-5 | - |
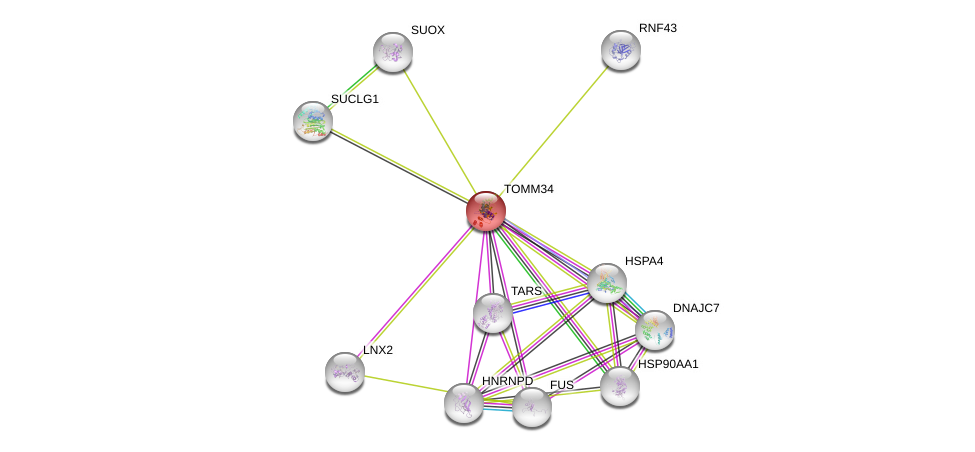
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005515 | protein binding | 21044950. | IPI | Function |
| GO:0005634 | nucleus | 21630459. | HDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005741 | mitochondrial outer membrane | 9324309. | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0006626 | protein targeting to mitochondrion | 9324309. | IMP | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016021 | integral component of membrane | 9324309. | TAS | Component |
| GO:0031072 | heat shock protein binding | 9660753. | IPI | Function |