
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| KIRC | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| PAAD | |||
| READ | |||
| SARC |
| Cancer | P-value | Q-value |
|---|---|---|
| SARC | ||
| KIRP | ||
| PAAD | ||
| BLCA | ||
| READ | ||
| KICH | ||
| UCEC | ||
| GBM | ||
| LIHC | ||
| DLBC | ||
| THCA | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000409921 | CAPG-205 | 1195 | - | ENSP00000387063 | 333 (aa) | - | P40121 |
| ENST00000479651 | CAPG-213 | 265 | - | - | - (aa) | - | - |
| ENST00000491267 | CAPG-215 | 549 | - | - | - (aa) | - | - |
| ENST00000471064 | CAPG-212 | 486 | - | - | - (aa) | - | - |
| ENST00000447219 | CAPG-208 | 930 | - | ENSP00000398232 | 253 (aa) | - | E7ENU9 |
| ENST00000409670 | CAPG-203 | 1183 | - | ENSP00000386315 | 348 (aa) | - | P40121 |
| ENST00000263867 | CAPG-201 | 1442 | XM_011533122 | ENSP00000263867 | 348 (aa) | XP_011531424 | P40121 |
| ENST00000449030 | CAPG-209 | 839 | - | ENSP00000403330 | 253 (aa) | - | E7ENU9 |
| ENST00000483659 | CAPG-214 | 767 | - | - | - (aa) | - | - |
| ENST00000439385 | CAPG-207 | 865 | - | ENSP00000391923 | 253 (aa) | - | E7ENU9 |
| ENST00000415012 | CAPG-206 | 531 | - | - | - (aa) | - | - |
| ENST00000409724 | CAPG-204 | 1151 | - | ENSP00000386965 | 348 (aa) | - | P40121 |
| ENST00000409275 | CAPG-202 | 835 | - | ENSP00000386596 | 218 (aa) | - | B8ZZL6 |
| ENST00000453973 | CAPG-210 | 452 | - | ENSP00000397381 | 103 (aa) | - | H7C0X8 |
| ENST00000459793 | CAPG-211 | 943 | - | - | - (aa) | - | - |
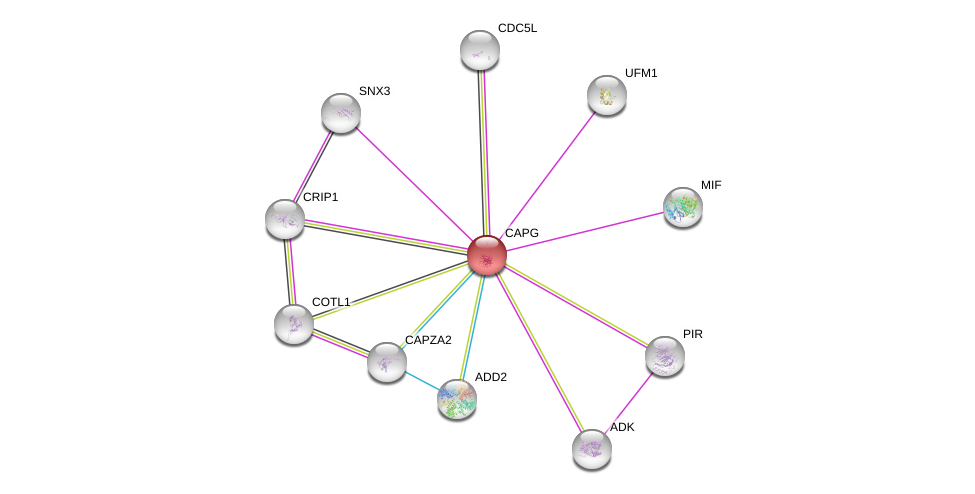
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000042493 | CAPG | 100 | 92.694 | ENSMUSG00000056737 | Capg | 100 | 92.550 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001726 | ruffle | - | IEA | Component |
| GO:0005515 | protein binding | 18266911.25416956. | IPI | Function |
| GO:0005634 | nucleus | 18266911. | IDA | Component |
| GO:0005654 | nucleoplasm | 18938132. | IDA | Component |
| GO:0005730 | nucleolus | 18938132. | IDA | Component |
| GO:0005737 | cytoplasm | 18266911.18938132.19166812.24236012.25468996. | IDA | Component |
| GO:0005814 | centriole | 19166812. | IDA | Component |
| GO:0008290 | F-actin capping protein complex | 1322908. | TAS | Component |
| GO:0019904 | protein domain specific binding | 18266911. | IPI | Function |
| GO:0022617 | extracellular matrix disassembly | 24236012. | IMP | Process |
| GO:0030027 | lamellipodium | - | IEA | Component |
| GO:0042470 | melanosome | - | IEA | Component |
| GO:0044877 | protein-containing complex binding | 18266911. | IDA | Function |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0051015 | actin filament binding | - | IEA | Function |
| GO:0051016 | barbed-end actin filament capping | - | IEA | Process |
| GO:0065003 | protein-containing complex assembly | 1322908. | NAS | Process |
| GO:0070062 | extracellular exosome | 19056867.20458337.23533145. | HDA | Component |
| GO:0071803 | positive regulation of podosome assembly | 24236012. | IMP | Process |
| GO:0072686 | mitotic spindle | 19166812. | IDA | Component |
| GO:0090543 | Flemming body | 19166812. | IDA | Component |