
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| KIRP | |||
| LAML | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| SKCM | |||
| THCA | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| BRCA | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| READ | ||
| KICH | ||
| LIHC |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000522013 | CTNNA1-232 | 874 | - | ENSP00000430379 | 105 (aa) | - | E5RFK9 |
| ENST00000517534 | CTNNA1-203 | 416 | - | - | - (aa) | - | - |
| ENST00000521640 | CTNNA1-228 | 568 | - | ENSP00000430623 | 40 (aa) | - | E5RJ43 |
| ENST00000521387 | CTNNA1-227 | 871 | - | - | - (aa) | - | - |
| ENST00000517533 | CTNNA1-202 | 579 | - | ENSP00000431118 | 119 (aa) | - | E5RJP7 |
| ENST00000521683 | CTNNA1-229 | 565 | - | ENSP00000430981 | 71 (aa) | - | E5RGU3 |
| ENST00000519113 | CTNNA1-213 | 859 | - | ENSP00000430078 | 139 (aa) | - | E5RGY6 |
| ENST00000518585 | CTNNA1-209 | 568 | - | - | - (aa) | - | - |
| ENST00000519116 | CTNNA1-214 | 562 | - | ENSP00000428894 | 91 (aa) | - | E5RFG3 |
| ENST00000521724 | CTNNA1-230 | 2741 | - | ENSP00000431033 | 37 (aa) | - | E5RJZ2 |
| ENST00000524292 | CTNNA1-242 | 557 | - | - | - (aa) | - | - |
| ENST00000519489 | CTNNA1-216 | 358 | - | - | - (aa) | - | - |
| ENST00000519634 | CTNNA1-217 | 589 | - | ENSP00000428088 | 50 (aa) | - | E5RGS1 |
| ENST00000302763 | CTNNA1-201 | 3756 | - | ENSP00000304669 | 906 (aa) | - | P35221 |
| ENST00000519768 | CTNNA1-218 | 560 | - | ENSP00000429103 | 20 (aa) | - | E5RJC9 |
| ENST00000520158 | CTNNA1-219 | 719 | - | ENSP00000429457 | 215 (aa) | - | E5RIB1 |
| ENST00000520339 | CTNNA1-221 | 868 | - | ENSP00000428202 | 101 (aa) | - | E5RJL0 |
| ENST00000524127 | CTNNA1-241 | 483 | - | ENSP00000428049 | 94 (aa) | - | E5RFM3 |
| ENST00000517904 | CTNNA1-205 | 661 | - | - | - (aa) | - | - |
| ENST00000520400 | CTNNA1-222 | 589 | - | - | - (aa) | - | - |
| ENST00000520520 | CTNNA1-223 | 484 | - | ENSP00000430076 | 136 (aa) | - | E5RGY7 |
| ENST00000520522 | CTNNA1-224 | 390 | - | ENSP00000428710 | 50 (aa) | - | E5RGS1 |
| ENST00000523685 | CTNNA1-239 | 694 | - | ENSP00000430240 | 128 (aa) | - | E5RGD2 |
| ENST00000518825 | CTNNA1-210 | 3536 | - | ENSP00000427821 | 841 (aa) | - | G3XAM7 |
| ENST00000523298 | CTNNA1-238 | 577 | - | ENSP00000428044 | 110 (aa) | - | E5RFM5 |
| ENST00000517980 | CTNNA1-206 | 696 | - | ENSP00000428439 | 117 (aa) | - | E5RIE0 |
| ENST00000627109 | CTNNA1-244 | 3831 | - | ENSP00000486200 | 841 (aa) | - | G3XAM7 |
| ENST00000518263 | CTNNA1-207 | 548 | - | - | - (aa) | - | - |
| ENST00000517656 | CTNNA1-204 | 554 | - | ENSP00000430177 | 39 (aa) | - | E5RGG4 |
| ENST00000519309 | CTNNA1-215 | 869 | - | ENSP00000430671 | 55 (aa) | - | E5RIT8 |
| ENST00000521368 | CTNNA1-226 | 544 | - | ENSP00000428686 | 22 (aa) | - | H0YB54 |
| ENST00000540387 | CTNNA1-243 | 2684 | - | ENSP00000438476 | 536 (aa) | - | P35221 |
| ENST00000520865 | CTNNA1-225 | 557 | - | ENSP00000430841 | 36 (aa) | - | E5RHR7 |
| ENST00000523275 | CTNNA1-237 | 536 | - | ENSP00000429142 | 129 (aa) | - | H0YBB8 |
| ENST00000522227 | CTNNA1-234 | 642 | - | ENSP00000429636 | 123 (aa) | - | E5RHV7 |
| ENST00000521941 | CTNNA1-231 | 224 | - | - | - (aa) | - | - |
| ENST00000518910 | CTNNA1-211 | 572 | - | ENSP00000430626 | 161 (aa) | - | E5RJ41 |
| ENST00000518381 | CTNNA1-208 | 487 | - | ENSP00000429738 | 75 (aa) | - | E5RHJ5 |
| ENST00000522792 | CTNNA1-236 | 2044 | - | - | - (aa) | - | - |
| ENST00000518919 | CTNNA1-212 | 498 | - | - | - (aa) | - | - |
| ENST00000522730 | CTNNA1-235 | 750 | - | - | - (aa) | - | - |
| ENST00000523912 | CTNNA1-240 | 545 | - | ENSP00000430304 | 156 (aa) | - | E5RG03 |
| ENST00000522052 | CTNNA1-233 | 579 | - | ENSP00000482738 | 64 (aa) | - | A0A087WZL6 |
| ENST00000520260 | CTNNA1-220 | 710 | - | ENSP00000429569 | 32 (aa) | - | E5RHY5 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04390 | Hippo signaling pathway | KEGG |
| hsa04520 | Adherens junction | KEGG |
| hsa04670 | Leukocyte transendothelial migration | KEGG |
| hsa05100 | Bacterial invasion of epithelial cells | KEGG |
| hsa05200 | Pathways in cancer | KEGG |
| hsa05213 | Endometrial cancer | KEGG |
| hsa05226 | Gastric cancer | KEGG |
| hsa05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000044115 | rs10043722 | 5 | 138901252 | A | General risk tolerance (MTAG) | 30643258 | [0.0059-0.0107] unit decrease | 0.0083 | EFO_0008579 |
| ENSG00000044115 | rs114916656 | 5 | 138671837 | T | General risk tolerance (MTAG) | 30643258 | [0.0079-0.0165] unit decrease | 0.0122 | EFO_0008579 |
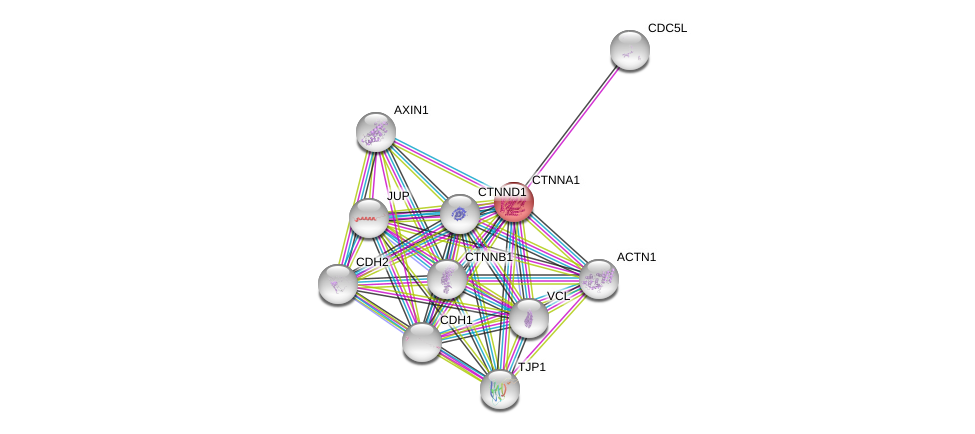
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000044115 | CTNNA1 | 95 | 32.075 | ENSG00000035403 | VCL | 54 | 38.614 | Homo_sapiens |
| ENSG00000044115 | CTNNA1 | 100 | 100.000 | ENSMUSG00000037815 | Ctnna1 | 100 | 99.448 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001541 | ovarian follicle development | - | IEA | Process |
| GO:0001669 | acrosomal vesicle | - | IEA | Component |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0005198 | structural molecule activity | - | IEA | Function |
| GO:0005515 | protein binding | 12477722.15657067.16212417.17098867.17620599.18093941.19016843.20936779.22750944.23292143.24189400.24509793.25241761.25764135.26496610.28051089. | IPI | Function |
| GO:0005794 | Golgi apparatus | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005886 | plasma membrane | - | IDA | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0005911 | cell-cell junction | 7650039.10460003. | IDA | Component |
| GO:0005915 | zonula adherens | - | IEA | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0007015 | actin filament organization | - | IEA | Process |
| GO:0007155 | cell adhesion | 8323564. | NAS | Process |
| GO:0007163 | establishment or maintenance of cell polarity | - | IEA | Process |
| GO:0007406 | negative regulation of neuroblast proliferation | - | IEA | Process |
| GO:0007568 | aging | - | IEA | Process |
| GO:0008013 | beta-catenin binding | 7650039.7890674. | IPI | Function |
| GO:0008584 | male gonad development | - | IEA | Process |
| GO:0014704 | intercalated disc | - | IEA | Component |
| GO:0015629 | actin cytoskeleton | - | IEA | Component |
| GO:0016264 | gap junction assembly | - | IEA | Process |
| GO:0016342 | catenin complex | 7650039.28051089. | IDA | Component |
| GO:0016600 | flotillin complex | - | IEA | Component |
| GO:0017166 | vinculin binding | 9700171. | IPI | Function |
| GO:0030027 | lamellipodium | - | IEA | Component |
| GO:0030054 | cell junction | - | IDA | Component |
| GO:0031103 | axon regeneration | - | IEA | Process |
| GO:0034332 | adherens junction organization | - | TAS | Process |
| GO:0034613 | cellular protein localization | - | IEA | Process |
| GO:0042475 | odontogenesis of dentin-containing tooth | - | IEA | Process |
| GO:0042802 | identical protein binding | 23292143. | IPI | Function |
| GO:0043231 | intracellular membrane-bounded organelle | - | IDA | Component |
| GO:0043297 | apical junction assembly | 9700171. | NAS | Process |
| GO:0043627 | response to estrogen | - | IEA | Process |
| GO:0045295 | gamma-catenin binding | 7650039.7890674. | IPI | Function |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0045296 | cadherin binding | 7650039.12734196. | IPI | Function |
| GO:0045880 | positive regulation of smoothened signaling pathway | - | IEA | Process |
| GO:0046982 | protein heterodimerization activity | - | IEA | Function |
| GO:0051015 | actin filament binding | - | IEA | Function |
| GO:0051149 | positive regulation of muscle cell differentiation | - | TAS | Process |
| GO:0051291 | protein heterooligomerization | - | IEA | Process |
| GO:0071681 | cellular response to indole-3-methanol | 10868478. | IDA | Process |
| GO:0090136 | epithelial cell-cell adhesion | - | IEA | Process |
| GO:2000146 | negative regulation of cell motility | - | IEA | Process |
| GO:2001045 | negative regulation of integrin-mediated signaling pathway | - | IEA | Process |
| GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | - | IEA | Process |
| GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand | - | IEA | Process |