
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| THCA | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CHOL | |||
| COAD | |||
| MESO | |||
| PAAD | |||
| READ | |||
| THCA | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| SARC | ||
| MESO | ||
| ACC | ||
| LUSC | ||
| BRCA | ||
| KIRP | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| LUAD |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000489386 | ALDH18A1-205 | 543 | - | - | - (aa) | - | - |
| ENST00000371221 | ALDH18A1-201 | 3337 | XM_024448096 | ENSP00000360265 | 793 (aa) | XP_024303864 | P54886 |
| ENST00000485428 | ALDH18A1-204 | 726 | - | - | - (aa) | - | - |
| ENST00000371224 | ALDH18A1-202 | 3359 | XM_024448094 | ENSP00000360268 | 795 (aa) | XP_024303862 | P54886 |
| ENST00000483788 | ALDH18A1-203 | 750 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000059573 | Metabolism | 8E-11 | 24816252 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000059573 | rs56322409 | 10 | 95636205 | T | Blood metabolite levels | 24816252 | [0.0071-0.0149] unit decrease | 0.011 | EFO_0005664 |
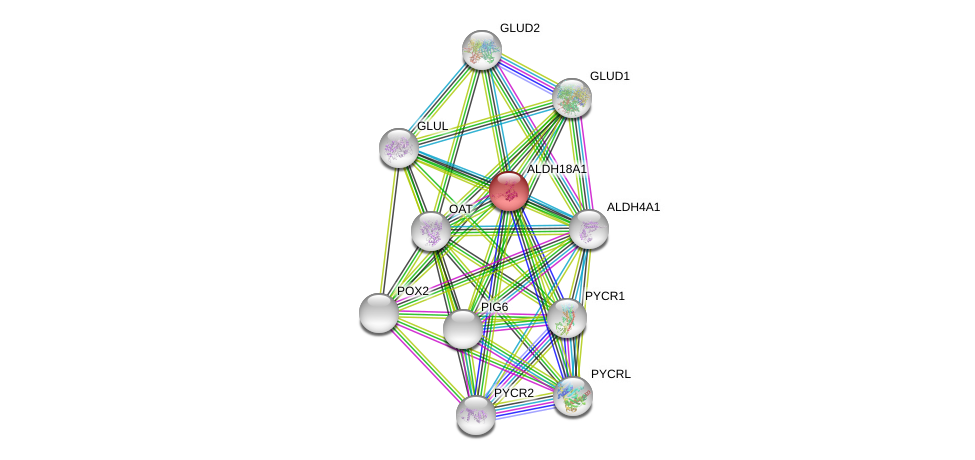
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000059573 | ALDH18A1 | 91 | 55.372 | WBGene00011938 | alh-13 | 89 | 55.372 | Caenorhabditis_elegans |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0004349 | glutamate 5-kinase activity | 11092761. | IDA | Function |
| GO:0004349 | glutamate 5-kinase activity | 26297558. | IMP | Function |
| GO:0004350 | glutamate-5-semialdehyde dehydrogenase activity | 21873635. | IBA | Function |
| GO:0004350 | glutamate-5-semialdehyde dehydrogenase activity | 11092761. | IDA | Function |
| GO:0004350 | glutamate-5-semialdehyde dehydrogenase activity | 26297558. | IMP | Function |
| GO:0005515 | protein binding | 25416956. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005739 | mitochondrion | 21873635. | IBA | Component |
| GO:0005739 | mitochondrion | 26297558.26320891. | IDA | Component |
| GO:0005743 | mitochondrial inner membrane | - | TAS | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0006536 | glutamate metabolic process | 11092761. | IMP | Process |
| GO:0006561 | proline biosynthetic process | 11092761.26297558. | IMP | Process |
| GO:0006592 | ornithine biosynthetic process | 11092761.26297558. | IMP | Process |
| GO:0008652 | cellular amino acid biosynthetic process | - | TAS | Process |
| GO:0016310 | phosphorylation | - | IEA | Process |
| GO:0019240 | citrulline biosynthetic process | 11092761.26297558. | IMP | Process |
| GO:0042802 | identical protein binding | 26297558. | IDA | Function |
| GO:0055114 | oxidation-reduction process | - | IEA | Process |
| GO:0055129 | L-proline biosynthetic process | - | IEA | Process |