
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| COAD | |||
| DLBC | |||
| GBM | |||
| LUAD | |||
| MESO | |||
| PAAD | |||
| READ | |||
| STAD | |||
| TGCT | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| ACC | ||
| SKCM | ||
| ESCA | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| LAML | ||
| LGG | ||
| LUAD |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000279550 | CSD | PF00313.22 | 1.6e-25 | 1 | 1 |
| ENSP00000228251 | CSD | PF00313.22 | 1.9e-25 | 1 | 1 |
| ENSP00000485718 | CSD | PF00313.22 | 4.1e-14 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000366290 | YBX3-204 | 512 | - | - | - (aa) | - | - |
| ENST00000366286 | YBX3-203 | 704 | - | - | - (aa) | - | - |
| ENST00000540447 | YBX3-208 | 6393 | - | - | - (aa) | - | - |
| ENST00000544622 | YBX3-216 | 685 | XM_011520871 | ENSP00000485718 | 92 (aa) | XP_011519173 | A0A0D9SEI8 |
| ENST00000228251 | YBX3-201 | 1796 | XM_017020122 | ENSP00000228251 | 372 (aa) | XP_016875611 | P16989 |
| ENST00000542002 | YBX3-212 | 952 | - | - | - (aa) | - | - |
| ENST00000546164 | YBX3-217 | 805 | - | - | - (aa) | - | - |
| ENST00000536107 | YBX3-205 | 438 | - | - | - (aa) | - | - |
| ENST00000540975 | YBX3-210 | 677 | - | - | - (aa) | - | - |
| ENST00000541351 | YBX3-211 | 717 | - | - | - (aa) | - | - |
| ENST00000544501 | YBX3-214 | 181 | - | - | - (aa) | - | - |
| ENST00000536823 | YBX3-206 | 840 | - | - | - (aa) | - | - |
| ENST00000542641 | YBX3-213 | 669 | - | - | - (aa) | - | - |
| ENST00000539204 | YBX3-207 | 649 | - | - | - (aa) | - | - |
| ENST00000279550 | YBX3-202 | 1708 | XM_017020123 | ENSP00000279550 | 303 (aa) | XP_016875612 | P16989 |
| ENST00000540747 | YBX3-209 | 548 | - | - | - (aa) | - | - |
| ENST00000544504 | YBX3-215 | 583 | - | - | - (aa) | - | - |
| ENST00000546298 | YBX3-218 | 564 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04530 | Tight junction | KEGG |
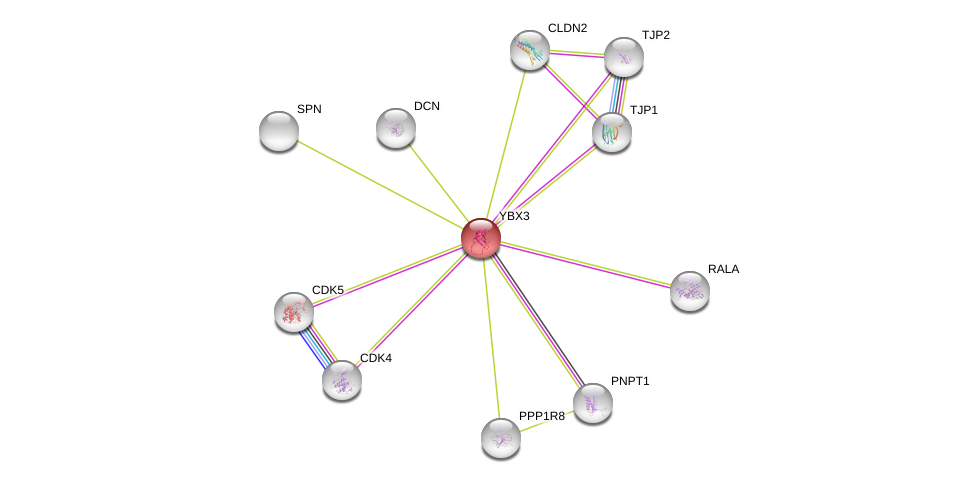
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000060138 | YBX3 | 70 | 95.312 | ENSG00000065978 | YBX1 | 85 | 55.034 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000060138 | YBX3 | 91 | 96.429 | ENSAMEG00000009393 | YBX3 | 95 | 90.000 | Ailuropoda_melanoleuca |
| ENSG00000060138 | YBX3 | 91 | 95.238 | ENSAPLG00000003147 | YBX3 | 70 | 100.000 | Anas_platyrhynchos |
| ENSG00000060138 | YBX3 | 98 | 86.923 | ENSANAG00000033366 | - | 93 | 86.923 | Aotus_nancymaae |
| ENSG00000060138 | YBX3 | 100 | 97.043 | ENSANAG00000024492 | - | 100 | 98.418 | Aotus_nancymaae |
| ENSG00000060138 | YBX3 | 89 | 100.000 | ENSBTAG00000009663 | YBX3 | 100 | 77.003 | Bos_taurus |
| ENSG00000060138 | YBX3 | 82 | 90.667 | ENSCJAG00000033824 | - | 98 | 91.892 | Callithrix_jacchus |
| ENSG00000060138 | YBX3 | 100 | 95.968 | ENSCJAG00000021660 | - | 100 | 95.968 | Callithrix_jacchus |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSCAFG00000013401 | YBX3 | 100 | 92.202 | Canis_familiaris |
| ENSG00000060138 | YBX3 | 100 | 87.467 | ENSCAFG00020006412 | YBX3 | 100 | 86.400 | Canis_lupus_dingo |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSCHIG00000010863 | YBX3 | 100 | 81.183 | Capra_hircus |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSTSYG00000004204 | - | 88 | 94.774 | Carlito_syrichta |
| ENSG00000060138 | YBX3 | 100 | 75.248 | ENSTSYG00000035821 | - | 97 | 80.357 | Carlito_syrichta |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSCAPG00000015039 | YBX3 | 93 | 86.760 | Cavia_aperea |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSCPOG00000011329 | YBX3 | 86 | 87.713 | Cavia_porcellus |
| ENSG00000060138 | YBX3 | 100 | 96.774 | ENSCCAG00000016469 | YBX3 | 100 | 96.774 | Cebus_capucinus |
| ENSG00000060138 | YBX3 | 100 | 98.660 | ENSCATG00000034411 | YBX3 | 100 | 98.660 | Cercocebus_atys |
| ENSG00000060138 | YBX3 | 91 | 96.429 | ENSCLAG00000016030 | YBX3 | 94 | 87.108 | Chinchilla_lanigera |
| ENSG00000060138 | YBX3 | 100 | 99.196 | ENSCSAG00000008744 | YBX3 | 100 | 99.196 | Chlorocebus_sabaeus |
| ENSG00000060138 | YBX3 | 89 | 100.000 | ENSCHOG00000011988 | YBX3 | 77 | 83.237 | Choloepus_hoffmanni |
| ENSG00000060138 | YBX3 | 92 | 97.647 | ENSCPBG00000014147 | YBX3 | 81 | 78.840 | Chrysemys_picta_bellii |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSCANG00000036801 | YBX3 | 100 | 98.413 | Colobus_angolensis_palliatus |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSCGRG00001016148 | Ybx3 | 100 | 84.409 | Cricetulus_griseus_chok1gshd |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSCGRG00000004563 | Ybx3 | 98 | 89.384 | Cricetulus_griseus_crigri |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSDORG00000009103 | Ybx3 | 81 | 90.592 | Dipodomys_ordii |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSETEG00000002557 | YBX3 | 99 | 84.155 | Echinops_telfairi |
| ENSG00000060138 | YBX3 | 100 | 86.022 | ENSEASG00005014726 | YBX3 | 97 | 88.764 | Equus_asinus_asinus |
| ENSG00000060138 | YBX3 | 100 | 88.978 | ENSECAG00000013777 | YBX3 | 100 | 89.516 | Equus_caballus |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSEEUG00000010415 | YBX3 | 100 | 92.254 | Erinaceus_europaeus |
| ENSG00000060138 | YBX3 | 100 | 92.246 | ENSFCAG00000027748 | YBX3 | 100 | 93.770 | Felis_catus |
| ENSG00000060138 | YBX3 | 97 | 89.888 | ENSFALG00000003661 | YBX3 | 71 | 98.837 | Ficedula_albicollis |
| ENSG00000060138 | YBX3 | 91 | 96.429 | ENSFDAG00000021192 | YBX3 | 92 | 84.321 | Fukomys_damarensis |
| ENSG00000060138 | YBX3 | 89 | 97.561 | ENSGALG00000031890 | YBX3 | 70 | 97.849 | Gallus_gallus |
| ENSG00000060138 | YBX3 | 92 | 97.647 | ENSGAGG00000017780 | YBX3 | 77 | 82.960 | Gopherus_agassizii |
| ENSG00000060138 | YBX3 | 100 | 99.732 | ENSGGOG00000001352 | YBX3 | 100 | 99.732 | Gorilla_gorilla |
| ENSG00000060138 | YBX3 | 91 | 94.048 | ENSHGLG00000014798 | - | 86 | 76.431 | Heterocephalus_glaber_female |
| ENSG00000060138 | YBX3 | 88 | 100.000 | ENSHGLG00000015495 | - | 80 | 76.364 | Heterocephalus_glaber_female |
| ENSG00000060138 | YBX3 | 82 | 100.000 | ENSHGLG00100003589 | YBX3 | 87 | 88.532 | Heterocephalus_glaber_male |
| ENSG00000060138 | YBX3 | 91 | 92.857 | ENSSTOG00000032577 | - | 100 | 81.720 | Ictidomys_tridecemlineatus |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSSTOG00000002108 | - | 88 | 86.411 | Ictidomys_tridecemlineatus |
| ENSG00000060138 | YBX3 | 86 | 64.198 | ENSJJAG00000014239 | Ybx3 | 96 | 85.227 | Jaculus_jaculus |
| ENSG00000060138 | YBX3 | 100 | 91.398 | ENSLAFG00000017247 | YBX3 | 100 | 91.935 | Loxodonta_africana |
| ENSG00000060138 | YBX3 | 100 | 98.925 | ENSMFAG00000039369 | YBX3 | 100 | 98.925 | Macaca_fascicularis |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSMMUG00000022469 | YBX3 | 100 | 98.769 | Macaca_mulatta |
| ENSG00000060138 | YBX3 | 100 | 98.656 | ENSMNEG00000045109 | YBX3 | 100 | 98.656 | Macaca_nemestrina |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSMLEG00000033618 | YBX3 | 79 | 98.606 | Mandrillus_leucophaeus |
| ENSG00000060138 | YBX3 | 91 | 96.429 | ENSMGAG00000014224 | YBX3 | 93 | 100.000 | Meleagris_gallopavo |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSMAUG00000012185 | Ybx3 | 98 | 86.293 | Mesocricetus_auratus |
| ENSG00000060138 | YBX3 | 89 | 97.561 | ENSMICG00000031459 | - | 100 | 69.967 | Microcebus_murinus |
| ENSG00000060138 | YBX3 | 100 | 90.591 | ENSMICG00000038002 | - | 100 | 92.204 | Microcebus_murinus |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSMOCG00000016291 | Ybx3 | 96 | 91.186 | Microtus_ochrogaster |
| ENSG00000060138 | YBX3 | 89 | 97.561 | ENSMODG00000018248 | YBX3 | 91 | 84.028 | Monodelphis_domestica |
| ENSG00000060138 | YBX3 | 91 | 97.619 | MGP_CAROLIEiJ_G0028947 | Ybx3 | 100 | 83.871 | Mus_caroli |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSMUSG00000030189 | Ybx3 | 100 | 83.871 | Mus_musculus |
| ENSG00000060138 | YBX3 | 91 | 97.619 | MGP_PahariEiJ_G0022709 | Ybx3 | 100 | 84.409 | Mus_pahari |
| ENSG00000060138 | YBX3 | 91 | 97.619 | MGP_SPRETEiJ_G0029994 | Ybx3 | 100 | 84.140 | Mus_spretus |
| ENSG00000060138 | YBX3 | 100 | 79.790 | ENSMPUG00000011810 | YBX3 | 100 | 79.265 | Mustela_putorius_furo |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSMLUG00000007476 | YBX3 | 100 | 91.450 | Myotis_lucifugus |
| ENSG00000060138 | YBX3 | 91 | 96.429 | ENSNGAG00000000710 | Ybx3 | 91 | 83.333 | Nannospalax_galili |
| ENSG00000060138 | YBX3 | 95 | 94.085 | ENSNLEG00000034844 | YBX3 | 100 | 94.085 | Nomascus_leucogenys |
| ENSG00000060138 | YBX3 | 77 | 97.183 | ENSMEUG00000002507 | - | 81 | 76.879 | Notamacropus_eugenii |
| ENSG00000060138 | YBX3 | 82 | 80.921 | ENSODEG00000017957 | YBX3 | 94 | 81.620 | Octodon_degus |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSOANG00000001663 | - | 79 | 97.170 | Ornithorhynchus_anatinus |
| ENSG00000060138 | YBX3 | 91 | 96.429 | ENSOCUG00000001889 | YBX3 | 85 | 87.119 | Oryctolagus_cuniculus |
| ENSG00000060138 | YBX3 | 100 | 89.974 | ENSOGAG00000013245 | YBX3 | 100 | 89.710 | Otolemur_garnettii |
| ENSG00000060138 | YBX3 | 89 | 100.000 | ENSOARG00000020919 | YBX3 | 99 | 82.492 | Ovis_aries |
| ENSG00000060138 | YBX3 | 100 | 92.742 | ENSPPAG00000039068 | YBX3 | 100 | 92.742 | Pan_paniscus |
| ENSG00000060138 | YBX3 | 100 | 91.979 | ENSPPRG00000005859 | YBX3 | 100 | 93.443 | Panthera_pardus |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSPTIG00000005322 | YBX3 | 81 | 92.334 | Panthera_tigris_altaica |
| ENSG00000060138 | YBX3 | 100 | 99.465 | ENSPTRG00000004681 | YBX3 | 100 | 99.465 | Pan_troglodytes |
| ENSG00000060138 | YBX3 | 100 | 98.925 | ENSPANG00000010280 | YBX3 | 100 | 98.925 | Papio_anubis |
| ENSG00000060138 | YBX3 | 92 | 96.471 | ENSPSIG00000009247 | YBX3 | 95 | 100.000 | Pelodiscus_sinensis |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSPEMG00000012132 | - | 99 | 87.227 | Peromyscus_maniculatus_bairdii |
| ENSG00000060138 | YBX3 | 93 | 95.349 | ENSPEMG00000010391 | - | 85 | 91.150 | Peromyscus_maniculatus_bairdii |
| ENSG00000060138 | YBX3 | 85 | 97.436 | ENSPCIG00000029949 | YBX3 | 100 | 79.615 | Phascolarctos_cinereus |
| ENSG00000060138 | YBX3 | 100 | 99.462 | ENSPPYG00000004279 | YBX3 | 100 | 99.462 | Pongo_abelii |
| ENSG00000060138 | YBX3 | 91 | 96.429 | ENSPCAG00000001397 | YBX3 | 95 | 100.000 | Procavia_capensis |
| ENSG00000060138 | YBX3 | 79 | 97.260 | ENSPCOG00000011663 | YBX3 | 100 | 85.769 | Propithecus_coquereli |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSPVAG00000006740 | YBX3 | 77 | 91.724 | Pteropus_vampyrus |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSRNOG00000005480 | Ybx3 | 100 | 84.677 | Rattus_norvegicus |
| ENSG00000060138 | YBX3 | 100 | 98.118 | ENSRBIG00000039208 | YBX3 | 100 | 98.118 | Rhinopithecus_bieti |
| ENSG00000060138 | YBX3 | 100 | 98.387 | ENSRROG00000029638 | YBX3 | 100 | 98.387 | Rhinopithecus_roxellana |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSSBOG00000030573 | YBX3 | 87 | 95.593 | Saimiri_boliviensis_boliviensis |
| ENSG00000060138 | YBX3 | 89 | 97.561 | ENSSHAG00000003262 | YBX3 | 98 | 80.756 | Sarcophilus_harrisii |
| ENSG00000060138 | YBX3 | 89 | 97.590 | ENSSPUG00000009911 | - | 66 | 87.857 | Sphenodon_punctatus |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSSSCG00000000633 | YBX3 | 100 | 89.764 | Sus_scrofa |
| ENSG00000060138 | YBX3 | 98 | 90.000 | ENSTGUG00000012047 | YBX3 | 74 | 98.077 | Taeniopygia_guttata |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSTBEG00000012570 | YBX3 | 81 | 94.539 | Tupaia_belangeri |
| ENSG00000060138 | YBX3 | 91 | 97.619 | ENSTTRG00000008045 | YBX3 | 100 | 84.586 | Tursiops_truncatus |
| ENSG00000060138 | YBX3 | 75 | 94.203 | ENSUAMG00000016662 | YBX3 | 100 | 88.489 | Ursus_americanus |
| ENSG00000060138 | YBX3 | 91 | 96.429 | ENSUMAG00000016118 | YBX3 | 94 | 91.638 | Ursus_maritimus |
| ENSG00000060138 | YBX3 | 57 | 100.000 | ENSVPAG00000000876 | YBX3 | 100 | 72.535 | Vicugna_pacos |
| ENSG00000060138 | YBX3 | 79 | 97.260 | ENSVVUG00000004956 | YBX3 | 100 | 89.209 | Vulpes_vulpes |
| ENSG00000060138 | YBX3 | 82 | 93.333 | ENSXETG00000016239 | ybx3 | 90 | 68.142 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000122 | negative regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | - | IEA | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | - | ISA | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 19274049. | ISM | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 19274049. | NAS | Function |
| GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | - | IEA | Function |
| GO:0001701 | in utero embryonic development | - | IEA | Process |
| GO:0003690 | double-stranded DNA binding | 2977358. | TAS | Function |
| GO:0003700 | DNA-binding transcription factor activity | 8710501. | NAS | Function |
| GO:0003714 | transcription corepressor activity | 8710501. | TAS | Function |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003730 | mRNA 3'-UTR binding | 22711822. | ISS | Function |
| GO:0005515 | protein binding | 25593058. | IPI | Function |
| GO:0005634 | nucleus | 22711822. | ISS | Component |
| GO:0005737 | cytoplasm | 7628487. | TAS | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005923 | bicellular tight junction | 22711822. | ISS | Component |
| GO:0007283 | spermatogenesis | - | IEA | Process |
| GO:0008584 | male gonad development | - | IEA | Process |
| GO:0009409 | response to cold | 8710501. | TAS | Process |
| GO:0009566 | fertilization | - | IEA | Process |
| GO:0017048 | Rho GTPase binding | - | ISS | Function |
| GO:0046622 | positive regulation of organ growth | - | IEA | Process |
| GO:0048471 | perinuclear region of cytoplasm | 22711822. | ISS | Component |
| GO:0048642 | negative regulation of skeletal muscle tissue development | - | IEA | Process |
| GO:0060546 | negative regulation of necroptotic process | 22711822. | IMP | Process |
| GO:0070935 | 3'-UTR-mediated mRNA stabilization | 22711822. | IC | Process |
| GO:0071356 | cellular response to tumor necrosis factor | 22711822. | IMP | Process |
| GO:0071474 | cellular hyperosmotic response | 22711822. | IMP | Process |
| GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 22711822. | IMP | Process |
| GO:1905538 | polysome binding | - | IEA | Function |
| GO:2000767 | positive regulation of cytoplasmic translation | 22711822. | ISS | Process |