
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| LAML | |||
| LUAD | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| STAD | |||
| THCA | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| ACC | ||
| UCS | ||
| HNSC | ||
| COAD | ||
| PAAD | ||
| BLCA | ||
| READ | ||
| LAML | ||
| UCEC | ||
| LIHC | ||
| LGG |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000509616 | SLC12A2-207 | 580 | - | - | - (aa) | - | - |
| ENST00000504416 | SLC12A2-204 | 3653 | - | - | - (aa) | - | - |
| ENST00000343225 | SLC12A2-202 | 5509 | - | ENSP00000340878 | 1196 (aa) | - | P55011 |
| ENST00000509205 | SLC12A2-206 | 4263 | - | ENSP00000427109 | 1150 (aa) | - | G3XAL9 |
| ENST00000262461 | SLC12A2-201 | 6885 | XM_017009771 | ENSP00000262461 | 1212 (aa) | XP_016865260 | P55011 |
| ENST00000502849 | SLC12A2-203 | 489 | - | - | - (aa) | - | - |
| ENST00000628403 | SLC12A2-208 | 3617 | - | ENSP00000486323 | 1150 (aa) | - | G3XAL9 |
| ENST00000507791 | SLC12A2-205 | 555 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000064651 | Body Weights and Measures | 5.20889768949391E-6 | 17903300 |
| ENSG00000064651 | Body Weights and Measures | 4.32118025117837E-6 | 17903300 |
| ENSG00000064651 | Body Weights and Measures | 4.32051119738854E-6 | 17903300 |
| ENSG00000064651 | Body Weights and Measures | 3.64322895623924E-6 | 17903300 |
| ENSG00000064651 | Stroke | 8.3589458E-004 | - |
| ENSG00000064651 | Carcinoid Tumor | 2E-6 | 21139019 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000064651 | rs10089 | 5 | 128186851 | ? | Ileal carcinoids | 21139019 | [1.79-3.70] | 2.56 | EFO_0004243 |
| ENSG00000064651 | rs7734927 | 5 | 128118043 | A | Red cell distribution width | 28957414 | [0.1-0.12] unit increase | 0.112536 | EFO_0005192 |
| ENSG00000064651 | rs3101725 | 5 | 128188326 | C | Varicose veins | 30566020 | [0.11-0.18] unit decrease | 0.143 | HP_0002619 |
| ENSG00000064651 | rs185689480 | 5 | 128186065 | ? | Red cell distribution width | 30595370 | EFO_0005192 | ||
| ENSG00000064651 | rs2568928 | 5 | 128151911 | ? | Eosinophil counts | 30595370 | EFO_0004842 | ||
| ENSG00000064651 | rs2568928 | 5 | 128151911 | ? | Heel bone mineral density | 30595370 | EFO_0009270 | ||
| ENSG00000064651 | rs1112956 | 5 | 128098106 | G | Breast cancer | 29059683 | [0.018-0.047] unit decrease | 0.0328 | EFO_0000305 |
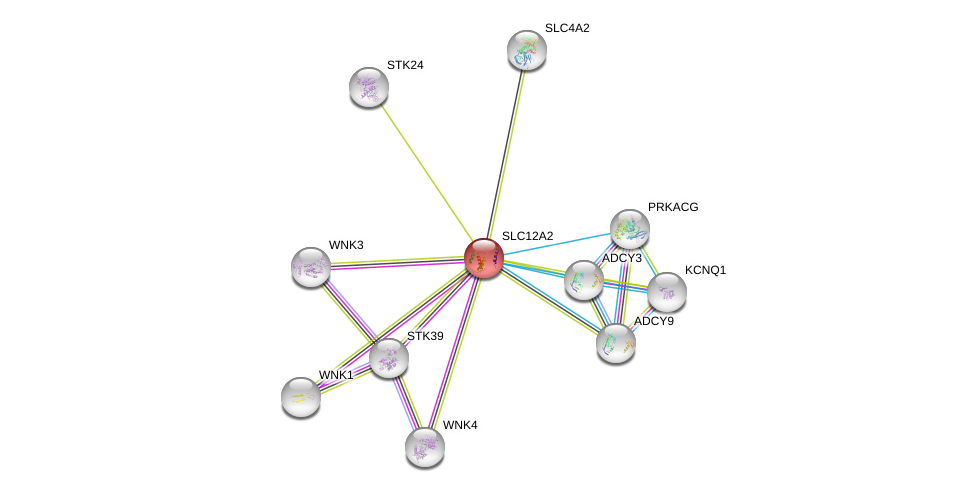
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005515 | protein binding | 17721439.22570591.24555568. | IPI | Function |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0005887 | integral component of plasma membrane | 7629105. | TAS | Component |
| GO:0006811 | ion transport | - | TAS | Process |
| GO:0006884 | cell volume homeostasis | 21873635. | IBA | Process |
| GO:0006972 | hyperosmotic response | - | IEA | Process |
| GO:0007214 | gamma-aminobutyric acid signaling pathway | - | IEA | Process |
| GO:0007568 | aging | - | IEA | Process |
| GO:0008511 | sodium:potassium:chloride symporter activity | 21873635. | IBA | Function |
| GO:0008519 | ammonium transmembrane transporter activity | 21873635. | IBA | Function |
| GO:0008519 | ammonium transmembrane transporter activity | 12946942.18032481. | IDA | Function |
| GO:0010818 | T cell chemotaxis | 27400149. | IMP | Process |
| GO:0015081 | sodium ion transmembrane transporter activity | 21873635. | IBA | Function |
| GO:0015378 | sodium:chloride symporter activity | 21873635. | IBA | Function |
| GO:0015379 | potassium:chloride symporter activity | 21873635. | IBA | Function |
| GO:0015696 | ammonium transport | 21873635. | IBA | Process |
| GO:0015696 | ammonium transport | 12946942.18032481. | IDA | Process |
| GO:0016020 | membrane | 7629105. | TAS | Component |
| GO:0016324 | apical plasma membrane | 21873635. | IBA | Component |
| GO:0019901 | protein kinase binding | 12740379.16669787. | IPI | Function |
| GO:0030321 | transepithelial chloride transport | 12946942. | IDA | Process |
| GO:0035725 | sodium ion transmembrane transport | 21873635. | IBA | Process |
| GO:0045795 | positive regulation of cell volume | - | IEA | Process |
| GO:0055064 | chloride ion homeostasis | 21873635. | IBA | Process |
| GO:0055075 | potassium ion homeostasis | 21873635. | IBA | Process |
| GO:0055078 | sodium ion homeostasis | 21873635. | IBA | Process |
| GO:0070062 | extracellular exosome | 19199708.20458337. | HDA | Component |
| GO:0070634 | transepithelial ammonium transport | 18032481. | IDA | Process |
| GO:0072488 | ammonium transmembrane transport | - | IEA | Process |
| GO:1902476 | chloride transmembrane transport | 21873635. | IBA | Process |
| GO:1903561 | extracellular vesicle | 24769233. | HDA | Component |
| GO:1990573 | potassium ion import across plasma membrane | 21873635. | IBA | Process |
| GO:1990869 | cellular response to chemokine | 27400149. | IMP | Process |