
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| MESO | ||
| BRCA | ||
| KIRP | ||
| COAD | ||
| BLCA | ||
| READ | ||
| LIHC | ||
| LUAD | ||
| OV |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000447013 | MMR_HSR1 | PF01926.23 | 2.6e-05 | 1 | 1 |
| ENSP00000266481 | MMR_HSR1 | PF01926.23 | 6.4e-05 | 1 | 1 |
| ENSP00000415131 | MMR_HSR1 | PF01926.23 | 6.6e-05 | 1 | 1 |
| ENSP00000448610 | MMR_HSR1 | PF01926.23 | 7.2e-05 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000547719 | DNM1L-214 | 601 | - | - | - (aa) | - | - |
| ENST00000546757 | DNM1L-209 | 2661 | - | ENSP00000448105 | 54 (aa) | - | H0YHY4 |
| ENST00000549926 | DNM1L-221 | 750 | - | - | - (aa) | - | - |
| ENST00000547640 | DNM1L-213 | 602 | - | - | - (aa) | - | - |
| ENST00000549701 | DNM1L-220 | 2487 | - | ENSP00000450399 | 736 (aa) | - | O00429 |
| ENST00000551643 | DNM1L-227 | 538 | - | ENSP00000450401 | 97 (aa) | - | F8VR28 |
| ENST00000266481 | DNM1L-201 | 3101 | XM_017018665 | ENSP00000266481 | 699 (aa) | XP_016874154 | O00429 |
| ENST00000551076 | DNM1L-225 | 556 | - | - | - (aa) | - | - |
| ENST00000548750 | DNM1L-218 | 813 | - | ENSP00000447788 | 261 (aa) | - | F8VZ52 |
| ENST00000548671 | DNM1L-217 | 750 | - | - | - (aa) | - | - |
| ENST00000358214 | DNM1L-202 | 3229 | - | ENSP00000350948 | 712 (aa) | - | G8JLD5 |
| ENST00000414834 | DNM1L-205 | 4211 | XM_011520544 | ENSP00000404160 | 533 (aa) | XP_011518846 | O00429 |
| ENST00000550093 | DNM1L-223 | 570 | - | - | - (aa) | - | - |
| ENST00000452533 | DNM1L-207 | 4439 | - | ENSP00000415131 | 710 (aa) | - | O00429 |
| ENST00000434676 | DNM1L-206 | 1008 | - | ENSP00000390090 | 156 (aa) | - | B4DPZ9 |
| ENST00000547078 | DNM1L-210 | 450 | - | ENSP00000448802 | 48 (aa) | - | H0YI79 |
| ENST00000552743 | DNM1L-228 | 606 | - | - | - (aa) | - | - |
| ENST00000547548 | DNM1L-212 | 724 | - | - | - (aa) | - | - |
| ENST00000548151 | DNM1L-216 | 557 | - | - | - (aa) | - | - |
| ENST00000547932 | DNM1L-215 | 417 | - | - | - (aa) | - | - |
| ENST00000550011 | DNM1L-222 | 576 | - | - | - (aa) | - | - |
| ENST00000381000 | DNM1L-203 | 4397 | - | ENSP00000370388 | 738 (aa) | - | O00429 |
| ENST00000549157 | DNM1L-219 | 556 | - | - | - (aa) | - | - |
| ENST00000546649 | DNM1L-208 | 2097 | - | ENSP00000448936 | 117 (aa) | - | F8VYL3 |
| ENST00000547312 | DNM1L-211 | 2393 | - | ENSP00000448610 | 725 (aa) | - | O00429 |
| ENST00000551476 | DNM1L-226 | 560 | - | ENSP00000447845 | 172 (aa) | - | F8VUJ9 |
| ENST00000553031 | DNM1L-229 | 755 | - | - | - (aa) | - | - |
| ENST00000413295 | DNM1L-204 | 1064 | - | ENSP00000396030 | 180 (aa) | - | B4DDQ3 |
| ENST00000553257 | DNM1L-230 | 2699 | XM_011520543 | ENSP00000449089 | 749 (aa) | XP_011518845 | O00429 |
| ENST00000550154 | DNM1L-224 | 550 | - | ENSP00000447013 | 168 (aa) | - | F8W1W3 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000087470 | Hemoglobin A2 | 7.871e-005 | - |
| ENSG00000087470 | Platelet Function Tests | 4.6500000E-006 | - |
| ENSG00000087470 | Platelet Function Tests | 4.8500000E-006 | - |
| ENSG00000087470 | Platelet Function Tests | 2.1100000E-006 | - |
| ENSG00000087470 | Platelet Function Tests | 4.3700000E-006 | - |
| ENSG00000087470 | Platelet Function Tests | 5.0700000E-006 | - |
| ENSG00000087470 | Platelet Function Tests | 4.6500000E-006 | - |
| ENSG00000087470 | Platelet Function Tests | 4.6500000E-006 | - |
| ENSG00000087470 | Platelet Function Tests | 4.6500000E-006 | - |
| ENSG00000087470 | Platelet Function Tests | 1.0425700E-005 | - |
| ENSG00000087470 | Platelet Function Tests | 1.6024100E-005 | - |
| ENSG00000087470 | Platelet Function Tests | 2.3203200E-005 | - |
| ENSG00000087470 | Platelet Function Tests | 1.0425700E-005 | - |
| ENSG00000087470 | Platelet Function Tests | 1.0425700E-005 | - |
| ENSG00000087470 | Platelet Function Tests | 4.6500000E-006 | - |
| ENSG00000087470 | Platelet Function Tests | 1.6531600E-005 | - |
| ENSG00000087470 | Platelet Function Tests | 1.0547200E-005 | - |
| ENSG00000087470 | Platelet Function Tests | 1.2399400E-005 | - |
| ENSG00000087470 | Platelet Function Tests | 5.7000000E-006 | - |
| ENSG00000087470 | Platelet Function Tests | 1.2991100E-005 | - |
| ENSG00000087470 | Platelet Function Tests | 5.4105233E-006 | - |
| ENSG00000087470 | Platelet Function Tests | 1.8905200E-005 | - |
| ENSG00000087470 | Platelet Function Tests | 1.8905200E-005 | - |
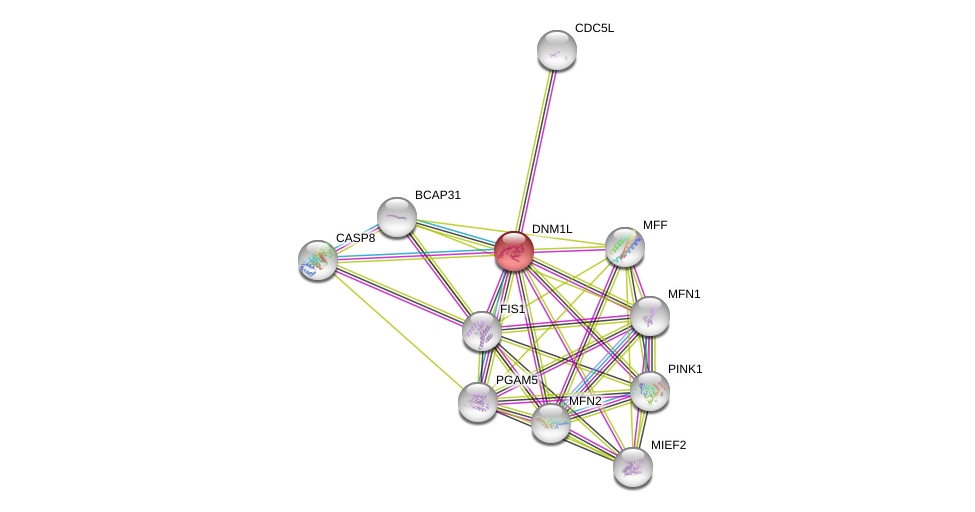
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000087470 | DNM1L | 100 | 87.286 | ENSAPOG00000022905 | - | 100 | 87.286 | Acanthochromis_polyacanthus |
| ENSG00000087470 | DNM1L | 100 | 87.692 | ENSAPOG00000016682 | dnm1l | 100 | 87.395 | Acanthochromis_polyacanthus |
| ENSG00000087470 | DNM1L | 100 | 87.518 | ENSACIG00000010115 | - | 100 | 87.571 | Amphilophus_citrinellus |
| ENSG00000087470 | DNM1L | 100 | 89.872 | ENSACIG00000002828 | dnm1l | 100 | 89.444 | Amphilophus_citrinellus |
| ENSG00000087470 | DNM1L | 100 | 87.429 | ENSAOCG00000005974 | - | 100 | 87.429 | Amphiprion_ocellaris |
| ENSG00000087470 | DNM1L | 100 | 88.604 | ENSAOCG00000010262 | dnm1l | 100 | 88.302 | Amphiprion_ocellaris |
| ENSG00000087470 | DNM1L | 100 | 87.552 | ENSAPEG00000005223 | dnm1l | 100 | 87.255 | Amphiprion_percula |
| ENSG00000087470 | DNM1L | 100 | 87.571 | ENSAPEG00000015504 | - | 100 | 87.268 | Amphiprion_percula |
| ENSG00000087470 | DNM1L | 100 | 87.268 | ENSATEG00000010274 | - | 100 | 93.519 | Anabas_testudineus |
| ENSG00000087470 | DNM1L | 100 | 87.732 | ENSATEG00000018248 | dnm1l | 100 | 87.732 | Anabas_testudineus |
| ENSG00000087470 | DNM1L | 100 | 100.000 | ENSANAG00000030575 | DNM1L | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000087470 | DNM1L | 100 | 86.695 | ENSACLG00000001401 | dnm1l | 100 | 86.695 | Astatotilapia_calliptera |
| ENSG00000087470 | DNM1L | 100 | 87.429 | ENSACLG00000003008 | - | 100 | 87.124 | Astatotilapia_calliptera |
| ENSG00000087470 | DNM1L | 100 | 90.028 | ENSAMXG00000001473 | dnm1l | 100 | 89.747 | Astyanax_mexicanus |
| ENSG00000087470 | DNM1L | 100 | 94.158 | ENSBTAG00000011395 | DNM1L | 100 | 94.158 | Bos_taurus |
| ENSG00000087470 | DNM1L | 100 | 94.429 | ENSCHIG00000024000 | DNM1L | 100 | 94.429 | Capra_hircus |
| ENSG00000087470 | DNM1L | 100 | 99.859 | ENSCCAG00000031176 | DNM1L | 100 | 99.859 | Cebus_capucinus |
| ENSG00000087470 | DNM1L | 100 | 100.000 | ENSCATG00000016013 | DNM1L | 100 | 100.000 | Cercocebus_atys |
| ENSG00000087470 | DNM1L | 100 | 99.864 | ENSCANG00000040112 | DNM1L | 100 | 99.864 | Colobus_angolensis_palliatus |
| ENSG00000087470 | DNM1L | 100 | 86.409 | ENSCSEG00000011310 | DNM1L | 100 | 86.123 | Cynoglossus_semilaevis |
| ENSG00000087470 | DNM1L | 98 | 84.314 | ENSCSEG00000014488 | dnm1l | 95 | 92.222 | Cynoglossus_semilaevis |
| ENSG00000087470 | DNM1L | 100 | 86.695 | ENSCVAG00000011641 | - | 100 | 86.552 | Cyprinodon_variegatus |
| ENSG00000087470 | DNM1L | 100 | 86.467 | ENSCVAG00000021070 | dnm1l | 100 | 86.838 | Cyprinodon_variegatus |
| ENSG00000087470 | DNM1L | 100 | 90.129 | ENSDARG00000015006 | dnm1l | 100 | 89.986 | Danio_rerio |
| ENSG00000087470 | DNM1L | 100 | 87.268 | ENSELUG00000015446 | DNM1L | 100 | 86.695 | Esox_lucius |
| ENSG00000087470 | DNM1L | 100 | 87.360 | ENSELUG00000009177 | dnm1l | 100 | 87.360 | Esox_lucius |
| ENSG00000087470 | DNM1L | 100 | 86.143 | ENSFHEG00000009442 | - | 95 | 92.593 | Fundulus_heteroclitus |
| ENSG00000087470 | DNM1L | 98 | 83.007 | ENSGMOG00000001805 | - | 100 | 81.960 | Gadus_morhua |
| ENSG00000087470 | DNM1L | 98 | 86.928 | ENSGMOG00000001033 | dnm1l | 99 | 83.911 | Gadus_morhua |
| ENSG00000087470 | DNM1L | 100 | 86.099 | ENSGAFG00000000857 | dnm1l | 100 | 86.448 | Gambusia_affinis |
| ENSG00000087470 | DNM1L | 100 | 86.467 | ENSGAFG00000017056 | - | 100 | 86.266 | Gambusia_affinis |
| ENSG00000087470 | DNM1L | 100 | 86.286 | ENSGACG00000018930 | - | 100 | 85.837 | Gasterosteus_aculeatus |
| ENSG00000087470 | DNM1L | 100 | 85.813 | ENSGACG00000004145 | dnm1l | 99 | 91.120 | Gasterosteus_aculeatus |
| ENSG00000087470 | DNM1L | 100 | 100.000 | ENSGGOG00000006636 | DNM1L | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000087470 | DNM1L | 98 | 88.235 | ENSHBUG00000004929 | - | 100 | 80.544 | Haplochromis_burtoni |
| ENSG00000087470 | DNM1L | 100 | 86.695 | ENSHBUG00000001952 | dnm1l | 100 | 86.695 | Haplochromis_burtoni |
| ENSG00000087470 | DNM1L | 98 | 89.003 | ENSIPUG00000006021 | dnm1l | 97 | 89.003 | Ictalurus_punctatus |
| ENSG00000087470 | DNM1L | 100 | 85.655 | ENSKMAG00000004994 | dnm1l | 100 | 85.655 | Kryptolebias_marmoratus |
| ENSG00000087470 | DNM1L | 98 | 86.928 | ENSKMAG00000021767 | - | 100 | 84.276 | Kryptolebias_marmoratus |
| ENSG00000087470 | DNM1L | 98 | 86.928 | ENSLBEG00000028335 | - | 97 | 91.346 | Labrus_bergylta |
| ENSG00000087470 | DNM1L | 100 | 87.255 | ENSLBEG00000008434 | dnm1l | 100 | 87.115 | Labrus_bergylta |
| ENSG00000087470 | DNM1L | 100 | 89.986 | ENSLACG00000009823 | DNM1L | 98 | 87.832 | Latimeria_chalumnae |
| ENSG00000087470 | DNM1L | 100 | 89.413 | ENSLOCG00000000949 | dnm1l | 100 | 89.413 | Lepisosteus_oculatus |
| ENSG00000087470 | DNM1L | 100 | 100.000 | ENSMFAG00000006058 | DNM1L | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000087470 | DNM1L | 100 | 100.000 | ENSMMUG00000013655 | DNM1L | 100 | 100.000 | Macaca_mulatta |
| ENSG00000087470 | DNM1L | 100 | 100.000 | ENSMNEG00000037323 | DNM1L | 100 | 100.000 | Macaca_nemestrina |
| ENSG00000087470 | DNM1L | 100 | 87.133 | ENSMAMG00000023186 | dnm1l | 100 | 86.975 | Mastacembelus_armatus |
| ENSG00000087470 | DNM1L | 100 | 87.983 | ENSMAMG00000006659 | - | 100 | 87.697 | Mastacembelus_armatus |
| ENSG00000087470 | DNM1L | 100 | 87.589 | ENSMZEG00005007753 | dnm1l | 100 | 87.589 | Maylandia_zebra |
| ENSG00000087470 | DNM1L | 98 | 85.621 | ENSMMOG00000002489 | - | 100 | 83.380 | Mola_mola |
| ENSG00000087470 | DNM1L | 98 | 87.582 | ENSMMOG00000014588 | dnm1l | 100 | 81.767 | Mola_mola |
| ENSG00000087470 | DNM1L | 100 | 96.603 | ENSMODG00000012763 | DNM1L | 100 | 96.603 | Monodelphis_domestica |
| ENSG00000087470 | DNM1L | 100 | 84.835 | ENSMALG00000003988 | - | 100 | 84.835 | Monopterus_albus |
| ENSG00000087470 | DNM1L | 100 | 87.411 | ENSMALG00000011851 | dnm1l | 100 | 87.268 | Monopterus_albus |
| ENSG00000087470 | DNM1L | 98 | 98.039 | ENSMUSG00000022789 | Dnm1l | 99 | 94.064 | Mus_musculus |
| ENSG00000087470 | DNM1L | 98 | 88.235 | ENSNBRG00000000347 | - | 100 | 83.192 | Neolamprologus_brichardi |
| ENSG00000087470 | DNM1L | 100 | 87.589 | ENSNBRG00000020969 | dnm1l | 100 | 87.589 | Neolamprologus_brichardi |
| ENSG00000087470 | DNM1L | 100 | 85.793 | ENSONIG00000008672 | dnm1l | 100 | 85.793 | Oreochromis_niloticus |
| ENSG00000087470 | DNM1L | 100 | 84.480 | ENSONIG00000016752 | - | 98 | 84.076 | Oreochromis_niloticus |
| ENSG00000087470 | DNM1L | 100 | 85.307 | ENSORLG00000003624 | dnm1l | 100 | 85.307 | Oryzias_latipes |
| ENSG00000087470 | DNM1L | 100 | 86.286 | ENSORLG00000010705 | - | 100 | 85.837 | Oryzias_latipes |
| ENSG00000087470 | DNM1L | 100 | 82.336 | ENSORLG00020002127 | - | 97 | 93.519 | Oryzias_latipes_hni |
| ENSG00000087470 | DNM1L | 100 | 85.449 | ENSORLG00020016524 | dnm1l | 100 | 85.449 | Oryzias_latipes_hni |
| ENSG00000087470 | DNM1L | 100 | 85.592 | ENSORLG00015009298 | dnm1l | 100 | 85.592 | Oryzias_latipes_hsok |
| ENSG00000087470 | DNM1L | 100 | 84.298 | ENSORLG00015001039 | - | 97 | 93.519 | Oryzias_latipes_hsok |
| ENSG00000087470 | DNM1L | 100 | 85.294 | ENSOMEG00000010635 | dnm1l | 100 | 85.294 | Oryzias_melastigma |
| ENSG00000087470 | DNM1L | 100 | 86.695 | ENSOMEG00000013872 | - | 100 | 86.409 | Oryzias_melastigma |
| ENSG00000087470 | DNM1L | 100 | 100.000 | ENSPPAG00000039981 | DNM1L | 100 | 100.000 | Pan_paniscus |
| ENSG00000087470 | DNM1L | 100 | 100.000 | ENSPTRG00000004817 | DNM1L | 100 | 100.000 | Pan_troglodytes |
| ENSG00000087470 | DNM1L | 100 | 88.062 | ENSPKIG00000001634 | dnm1l | 100 | 88.062 | Paramormyrops_kingsleyae |
| ENSG00000087470 | DNM1L | 98 | 86.928 | ENSPKIG00000010643 | dnm1l | 98 | 92.742 | Paramormyrops_kingsleyae |
| ENSG00000087470 | DNM1L | 98 | 80.745 | ENSPMGG00000019068 | - | 81 | 89.228 | Periophthalmus_magnuspinnatus |
| ENSG00000087470 | DNM1L | 100 | 85.408 | ENSPMGG00000005630 | dnm1l | 100 | 85.122 | Periophthalmus_magnuspinnatus |
| ENSG00000087470 | DNM1L | 100 | 93.057 | ENSPCIG00000015246 | DNM1L | 100 | 93.057 | Phascolarctos_cinereus |
| ENSG00000087470 | DNM1L | 98 | 86.928 | ENSPFOG00000002801 | dnm1l | 100 | 85.103 | Poecilia_formosa |
| ENSG00000087470 | DNM1L | 100 | 87.429 | ENSPFOG00000012922 | - | 100 | 87.124 | Poecilia_formosa |
| ENSG00000087470 | DNM1L | 100 | 86.667 | ENSPLAG00000001690 | dnm1l | 100 | 87.019 | Poecilia_latipinna |
| ENSG00000087470 | DNM1L | 100 | 87.571 | ENSPLAG00000012587 | DNM1L | 100 | 87.268 | Poecilia_latipinna |
| ENSG00000087470 | DNM1L | 100 | 87.429 | ENSPMEG00000021909 | - | 100 | 87.124 | Poecilia_mexicana |
| ENSG00000087470 | DNM1L | 100 | 86.667 | ENSPMEG00000006603 | dnm1l | 100 | 87.019 | Poecilia_mexicana |
| ENSG00000087470 | DNM1L | 100 | 85.551 | ENSPREG00000009857 | - | 100 | 85.122 | Poecilia_reticulata |
| ENSG00000087470 | DNM1L | 100 | 86.383 | ENSPREG00000008602 | dnm1l | 100 | 86.733 | Poecilia_reticulata |
| ENSG00000087470 | DNM1L | 100 | 87.554 | ENSPNYG00000006979 | - | 100 | 87.268 | Pundamilia_nyererei |
| ENSG00000087470 | DNM1L | 100 | 88.301 | ENSPNAG00000020017 | dnm1l | 100 | 88.000 | Pygocentrus_nattereri |
| ENSG00000087470 | DNM1L | 100 | 100.000 | ENSRBIG00000035947 | DNM1L | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000087470 | DNM1L | 100 | 100.000 | ENSRROG00000042126 | DNM1L | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000087470 | DNM1L | 100 | 99.718 | ENSSBOG00000026857 | DNM1L | 100 | 99.718 | Saimiri_boliviensis_boliviensis |
| ENSG00000087470 | DNM1L | 98 | 88.235 | ENSSFOG00015001002 | - | 100 | 84.406 | Scleropages_formosus |
| ENSG00000087470 | DNM1L | 100 | 89.270 | ENSSFOG00015004634 | dnm1l | 100 | 88.841 | Scleropages_formosus |
| ENSG00000087470 | DNM1L | 100 | 86.981 | ENSSMAG00000019922 | - | 100 | 86.695 | Scophthalmus_maximus |
| ENSG00000087470 | DNM1L | 100 | 85.734 | ENSSMAG00000008336 | dnm1l | 100 | 85.434 | Scophthalmus_maximus |
| ENSG00000087470 | DNM1L | 100 | 89.158 | ENSSDUG00000022859 | dnm1l | 100 | 89.143 | Seriola_dumerili |
| ENSG00000087470 | DNM1L | 100 | 87.714 | ENSSDUG00000008937 | - | 100 | 87.268 | Seriola_dumerili |
| ENSG00000087470 | DNM1L | 100 | 86.981 | ENSSLDG00000014797 | DNM1L | 100 | 86.838 | Seriola_lalandi_dorsalis |
| ENSG00000087470 | DNM1L | 100 | 87.857 | ENSSLDG00000006832 | - | 100 | 87.411 | Seriola_lalandi_dorsalis |
| ENSG00000087470 | DNM1L | 100 | 88.210 | ENSSPAG00000012049 | dnm1l | 100 | 88.352 | Stegastes_partitus |
| ENSG00000087470 | DNM1L | 100 | 86.838 | ENSSPAG00000015832 | - | 100 | 86.838 | Stegastes_partitus |
| ENSG00000087470 | DNM1L | 100 | 86.657 | ENSTRUG00000016834 | dnm1l | 100 | 86.657 | Takifugu_rubripes |
| ENSG00000087470 | DNM1L | 100 | 82.690 | ENSTNIG00000018526 | - | 100 | 82.546 | Tetraodon_nigroviridis |
| ENSG00000087470 | DNM1L | 98 | 92.810 | ENSXETG00000012252 | dnm1l | 100 | 91.866 | Xenopus_tropicalis |
| ENSG00000087470 | DNM1L | 100 | 87.714 | ENSXMAG00000011022 | - | 100 | 87.411 | Xiphophorus_maculatus |
| ENSG00000087470 | DNM1L | 100 | 85.794 | ENSXMAG00000017047 | dnm1l | 100 | 85.714 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000139 | Golgi membrane | - | IEA | Component |
| GO:0000266 | mitochondrial fission | 21873635. | IBA | Process |
| GO:0000266 | mitochondrial fission | 23530241. | IDA | Process |
| GO:0000266 | mitochondrial fission | 23349293.23921378.26992161.27145208. | IMP | Process |
| GO:0001836 | release of cytochrome c from mitochondria | 20850011. | IMP | Process |
| GO:0003374 | dynamin family protein polymerization involved in mitochondrial fission | 21873635. | IBA | Process |
| GO:0003374 | dynamin family protein polymerization involved in mitochondrial fission | 11514614.23530241. | IDA | Process |
| GO:0003924 | GTPase activity | 21873635. | IBA | Function |
| GO:0003924 | GTPase activity | 22265414.23530241. | IDA | Function |
| GO:0005096 | GTPase activator activity | 25767741. | IC | Function |
| GO:0005515 | protein binding | 9731200.12861026.18838687.19638400.21149567.21217774.21508961.21701560.22228096.22367970.22639965.23283981.23813973.24282027.24550280.26618722.27059175. | IPI | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005737 | cytoplasm | 12618434.21149567.23349293. | IDA | Component |
| GO:0005737 | cytoplasm | 28969390. | IMP | Component |
| GO:0005739 | mitochondrion | 11514614.22265414.23349293.23530241.23921378.24599962.26618722.29307555. | IDA | Component |
| GO:0005739 | mitochondrion | 28969390. | IMP | Component |
| GO:0005741 | mitochondrial outer membrane | 21149567.26122121.27145208. | IDA | Component |
| GO:0005741 | mitochondrial outer membrane | - | TAS | Component |
| GO:0005777 | peroxisome | 12618434. | IDA | Component |
| GO:0005777 | peroxisome | 23921378. | IMP | Component |
| GO:0005783 | endoplasmic reticulum | 9472031. | IDA | Component |
| GO:0005783 | endoplasmic reticulum | 11514614. | IDA | Component |
| GO:0005789 | endoplasmic reticulum membrane | 25767741. | TAS | Component |
| GO:0005794 | Golgi apparatus | 20688057. | IDA | Component |
| GO:0005829 | cytosol | 22265414.23921378. | IDA | Component |
| GO:0005829 | cytosol | 11514614. | IMP | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005874 | microtubule | 9472031. | IDA | Component |
| GO:0005903 | brush border | - | IEA | Component |
| GO:0005905 | clathrin-coated pit | - | IEA | Component |
| GO:0006816 | calcium ion transport | - | IEA | Process |
| GO:0007005 | mitochondrion organization | 11514614. | IMP | Process |
| GO:0007029 | endoplasmic reticulum organization | 11514614. | IMP | Process |
| GO:0008017 | microtubule binding | 21873635. | IBA | Function |
| GO:0008289 | lipid binding | - | IEA | Function |
| GO:0008637 | apoptotic mitochondrial changes | 21873635. | IBA | Process |
| GO:0010468 | regulation of gene expression | - | IEA | Process |
| GO:0010637 | negative regulation of mitochondrial fusion | - | IEA | Process |
| GO:0010821 | regulation of mitochondrion organization | 21873635. | IBA | Process |
| GO:0010821 | regulation of mitochondrion organization | 18353969.21149567. | IMP | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016020 | membrane | 21873635. | IBA | Component |
| GO:0016559 | peroxisome fission | 12618434. | IDA | Process |
| GO:0016559 | peroxisome fission | 18353969.23921378. | IMP | Process |
| GO:0017137 | Rab GTPase binding | 25767741. | IDA | Function |
| GO:0030054 | cell junction | - | IEA | Component |
| GO:0030276 | clathrin binding | - | IEA | Function |
| GO:0030672 | synaptic vesicle membrane | - | IEA | Component |
| GO:0030742 | GTP-dependent protein binding | 25767741. | IDA | Function |
| GO:0031625 | ubiquitin protein ligase binding | 16874301.19407830. | IPI | Function |
| GO:0031966 | mitochondrial membrane | 21873635. | IBA | Component |
| GO:0032459 | regulation of protein oligomerization | 20850011. | IDA | Process |
| GO:0032991 | protein-containing complex | 17408615. | IDA | Component |
| GO:0036466 | synaptic vesicle recycling via endosome | - | IEA | Process |
| GO:0042803 | protein homodimerization activity | 22265414.23530241. | IDA | Function |
| GO:0043065 | positive regulation of apoptotic process | 20850011. | IMP | Process |
| GO:0043231 | intracellular membrane-bounded organelle | - | IDA | Component |
| GO:0043547 | positive regulation of GTPase activity | - | IEA | Process |
| GO:0043653 | mitochondrial fragmentation involved in apoptotic process | 21873635. | IBA | Process |
| GO:0043653 | mitochondrial fragmentation involved in apoptotic process | 12499352.18353969.21149567. | IMP | Process |
| GO:0044877 | protein-containing complex binding | - | IEA | Function |
| GO:0048312 | intracellular distribution of mitochondria | 21873635. | IBA | Process |
| GO:0048312 | intracellular distribution of mitochondria | 11514614. | IMP | Process |
| GO:0048471 | perinuclear region of cytoplasm | 9472031.9570752. | IDA | Component |
| GO:0048488 | synaptic vesicle endocytosis | - | IEA | Process |
| GO:0050714 | positive regulation of protein secretion | 9570752. | IDA | Process |
| GO:0051259 | protein complex oligomerization | 27145208. | IMP | Process |
| GO:0051289 | protein homotetramerization | 18353969. | IDA | Process |
| GO:0051433 | BH2 domain binding | - | IEA | Function |
| GO:0060047 | heart contraction | - | IEA | Process |
| GO:0061003 | positive regulation of dendritic spine morphogenesis | - | IEA | Process |
| GO:0061025 | membrane fusion | 21873635. | IBA | Process |
| GO:0061025 | membrane fusion | 20850011. | IDA | Process |
| GO:0070266 | necroptotic process | 22265414. | IMP | Process |
| GO:0070584 | mitochondrion morphogenesis | 21149567. | IMP | Process |
| GO:0070585 | protein localization to mitochondrion | - | IEA | Process |
| GO:0090141 | positive regulation of mitochondrial fission | 19279012. | IMP | Process |
| GO:0090141 | positive regulation of mitochondrial fission | 20436456. | TAS | Process |
| GO:0090149 | mitochondrial membrane fission | 11514614. | IDA | Process |
| GO:0090149 | mitochondrial membrane fission | 27301544.27328748. | IMP | Process |
| GO:0090200 | positive regulation of release of cytochrome c from mitochondria | 21149567. | IMP | Process |
| GO:0090650 | cellular response to oxygen-glucose deprivation | - | IEA | Process |
| GO:0097194 | execution phase of apoptosis | - | TAS | Process |
| GO:0098794 | postsynapse | - | IEA | Component |
| GO:0098835 | presynaptic endocytic zone membrane | - | IEA | Component |
| GO:0099073 | mitochondrion-derived vesicle | 26618722. | IDA | Component |
| GO:1900063 | regulation of peroxisome organization | 18353969. | IMP | Process |
| GO:1900244 | positive regulation of synaptic vesicle endocytosis | - | IEA | Process |
| GO:1903146 | regulation of autophagy of mitochondrion | 21873635. | IBA | Process |
| GO:1903146 | regulation of autophagy of mitochondrion | 19279012. | IGI | Process |
| GO:1903578 | regulation of ATP metabolic process | - | IEA | Process |
| GO:1904579 | cellular response to thapsigargin | - | IEA | Process |
| GO:1904666 | regulation of ubiquitin protein ligase activity | - | IEA | Process |
| GO:1905395 | response to flavonoid | - | IEA | Process |
| GO:1990910 | response to hypobaric hypoxia | - | IEA | Process |
| GO:2000302 | positive regulation of synaptic vesicle exocytosis | - | IEA | Process |
| GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 20850011. | IMP | Process |