
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| COAD | |||
| DLBC | |||
| HNSC | |||
| LUAD | |||
| PAAD | |||
| SKCM | |||
| STAD | |||
| TGCT |
| Cancer | P-value | Q-value |
|---|---|---|
| MESO | ||
| ACC | ||
| LUSC | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| PCPG | ||
| BLCA | ||
| LAML | ||
| KICH | ||
| UCEC | ||
| GBM | ||
| LGG | ||
| OV |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 28900157 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000641756 | PHGDH-226 | 2132 | - | ENSP00000493147 | 70 (aa) | - | A0A286YEZ0 |
| ENST00000369407 | PHGDH-201 | 3180 | - | - | - (aa) | - | - |
| ENST00000641513 | PHGDH-219 | 735 | - | ENSP00000493398 | 165 (aa) | - | A0A286YFB2 |
| ENST00000641927 | PHGDH-230 | 1678 | - | - | - (aa) | - | - |
| ENST00000641587 | PHGDH-222 | 566 | - | ENSP00000493453 | 63 (aa) | - | A0A286YFF3 |
| ENST00000641947 | PHGDH-232 | 1774 | - | ENSP00000492994 | 526 (aa) | - | A0A286YF22 |
| ENST00000462324 | PHGDH-203 | 767 | - | - | - (aa) | - | - |
| ENST00000641023 | PHGDH-208 | 2096 | XM_011541226 | ENSP00000493175 | 533 (aa) | XP_011539528 | O43175 |
| ENST00000641213 | PHGDH-211 | 2053 | - | ENSP00000493079 | 165 (aa) | - | A0A286YFB2 |
| ENST00000641371 | PHGDH-215 | 725 | - | ENSP00000493305 | 145 (aa) | - | A0A286YFK5 |
| ENST00000641314 | PHGDH-214 | 1558 | - | - | - (aa) | - | - |
| ENST00000641074 | PHGDH-209 | 1780 | - | ENSP00000493446 | 429 (aa) | - | A0A286YFL2 |
| ENST00000641375 | PHGDH-216 | 2196 | - | ENSP00000493089 | 165 (aa) | - | A0A286YFB2 |
| ENST00000641272 | PHGDH-213 | 758 | - | ENSP00000493432 | 239 (aa) | - | A0A286YFM8 |
| ENST00000641720 | PHGDH-225 | 548 | - | ENSP00000492948 | 72 (aa) | - | A0A286YF34 |
| ENST00000641891 | PHGDH-229 | 2175 | - | ENSP00000493288 | 165 (aa) | - | A0A286YFB2 |
| ENST00000493622 | PHGDH-206 | 742 | - | ENSP00000493433 | 89 (aa) | - | A0A286YFE1 |
| ENST00000641847 | PHGDH-228 | 692 | - | - | - (aa) | - | - |
| ENST00000469443 | PHGDH-204 | 653 | - | - | - (aa) | - | - |
| ENST00000641939 | PHGDH-231 | 540 | - | - | - (aa) | - | - |
| ENST00000641811 | PHGDH-227 | 1031 | - | ENSP00000493296 | 276 (aa) | - | A0A286YF78 |
| ENST00000369409 | PHGDH-202 | 1882 | - | ENSP00000358417 | 532 (aa) | - | A0A2C9F2M7 |
| ENST00000641711 | PHGDH-224 | 712 | - | - | - (aa) | - | - |
| ENST00000496756 | PHGDH-207 | 508 | - | - | - (aa) | - | - |
| ENST00000642021 | PHGDH-233 | 2794 | - | - | - (aa) | - | - |
| ENST00000641115 | PHGDH-210 | 1638 | - | ENSP00000493264 | 445 (aa) | - | A0A286YFA2 |
| ENST00000482968 | PHGDH-205 | 1754 | - | - | - (aa) | - | - |
| ENST00000641491 | PHGDH-218 | 986 | - | ENSP00000493187 | 165 (aa) | - | A0A286YFB2 |
| ENST00000641597 | PHGDH-223 | 2585 | - | ENSP00000493382 | 533 (aa) | - | O43175 |
| ENST00000641455 | PHGDH-217 | 312 | - | - | - (aa) | - | - |
| ENST00000641573 | PHGDH-221 | 1180 | - | - | - (aa) | - | - |
| ENST00000641570 | PHGDH-220 | 1036 | - | ENSP00000493213 | 123 (aa) | - | A0A286YFC8 |
| ENST00000641247 | PHGDH-212 | 710 | - | ENSP00000492955 | 97 (aa) | - | A0A286YER3 |
| ENST00000642041 | PHGDH-234 | 943 | - | ENSP00000493415 | 97 (aa) | - | A0A286YER3 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000092621 | Vascular Diseases | 1.4600000E-005 | - |
| ENSG00000092621 | Glomerular Filtration Rate | 3.0081764E-005 | - |
| ENSG00000092621 | Metabolism | 3E-14 | 21886157 |
| ENSG00000092621 | Macular Degeneration | 2E-14 | 28250457 |
| ENSG00000092621 | Amino Acids | 3E-21 | 26068415 |
| ENSG00000092621 | Metabolism | 3E-21 | 26068415 |
| ENSG00000092621 | Insulin Resistance | 2E-14 | 23378610 |
| ENSG00000092621 | Body Height | 2E-9 | 25282103 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000092621 | rs523395 | 1 | 119729854 | C | Red blood cell count | 27863252 | [0.015-0.029] unit increase | 0.02186835 | EFO_0004305 |
| ENSG00000092621 | rs454510 | 1 | 119652419 | A | Alcohol dependence or chronic alcoholic pancreatitis or alcohol-related liver cirrhosis | 28714907 | 1.206 | EFO_1000802|EFO_0003829|EFO_1002013 | |
| ENSG00000092621 | rs478093 | 1 | 119712503 | G | Metabolite levels | 23378610 | EFO_0004471 | ||
| ENSG00000092621 | rs477992 | 1 | 119714953 | A | Amino acid levels | 26068415 | [0.031-0.048] unit decrease | 0.0397 | EFO_0005664|EFO_0005134 |
| ENSG00000092621 | rs1163251 | 1 | 119667132 | T | Blood metabolite levels | 24816252 | [0.015-0.023] unit increase | 0.019 | EFO_0005664 |
| ENSG00000092621 | rs666930 | 1 | 119716347 | G | Multiple sclerosis | 24076602 | 1.08 | EFO_0003885 | |
| ENSG00000092621 | rs72988658 | 1 | 119713339 | ? | Measles | 28928442 | [1.28-3.5] unit increase | 2.3883 | EFO_0008414 |
| ENSG00000092621 | rs477992 | 1 | 119714953 | A | Metabolic traits | 21886157 | [NR] unit decrease | 0.051 | EFO_0004725 |
| ENSG00000092621 | rs894079 | 1 | 119721034 | ? | Severe gingival inflammation | 28459102 | [0.29-0.68] unit increase | 0.48272 | HP_0000225 |
| ENSG00000092621 | rs12144094 | 1 | 119722200 | C | Height | 25282103 | [0.017-0.033] unit increase | 0.025 | EFO_0004339 |
| ENSG00000092621 | rs637868 | 1 | 119714487 | C | Alanine aminotransferase (ALT) levels after remission induction therapy in actute lymphoblastic leukemia (ALL) | 28090653 | [1.08-2.22] | 1.5384616 | EFO_0004735|EFO_0007965|EFO_0000220 |
| ENSG00000092621 | rs12072780 | 1 | 119685572 | T | Nicotine dependence and major depression (severity of comorbidity) | 30287806 | [0.35-0.86] unit increase | 0.6092 | EFO_0009262|EFO_0007006 |
| ENSG00000092621 | rs662602 | 1 | 119724280 | ? | Red blood cell count | 30595370 | EFO_0004305 | ||
| ENSG00000092621 | rs666930 | 1 | 119716347 | C | Breast cancer | 29059683 | [0.022-0.047] unit increase | 0.0347 | EFO_0000305 |
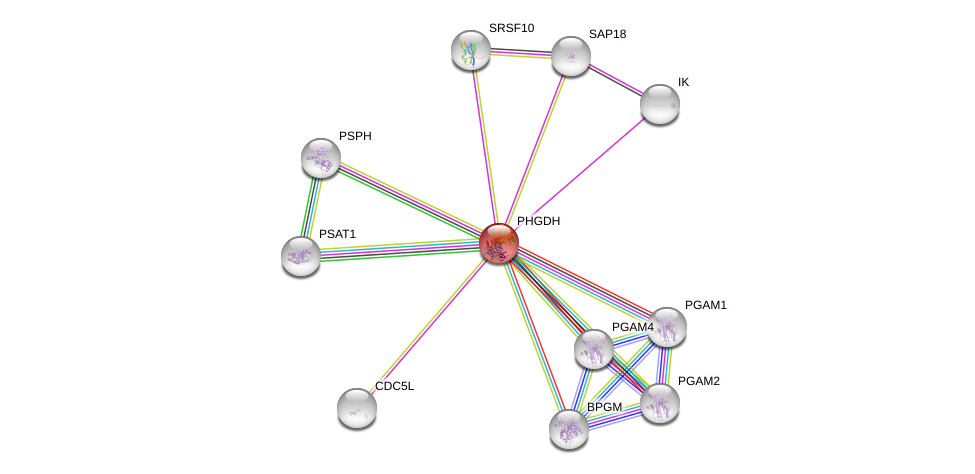
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000092621 | PHGDH | 94 | 63.095 | FBgn0032350 | CG6287 | 98 | 52.923 | Drosophila_melanogaster |
| ENSG00000092621 | PHGDH | 100 | 100.000 | ENSMUSG00000053398 | Phgdh | 100 | 94.559 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004617 | phosphoglycerate dehydrogenase activity | 21873635. | IBA | Function |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006520 | cellular amino acid metabolic process | 21873635. | IBA | Process |
| GO:0006541 | glutamine metabolic process | - | IEA | Process |
| GO:0006544 | glycine metabolic process | - | IEA | Process |
| GO:0006564 | L-serine biosynthetic process | - | TAS | Process |
| GO:0006566 | threonine metabolic process | - | IEA | Process |
| GO:0007420 | brain development | 8758134. | TAS | Process |
| GO:0009055 | electron transfer activity | 8758134. | TAS | Function |
| GO:0009448 | gamma-aminobutyric acid metabolic process | - | IEA | Process |
| GO:0010468 | regulation of gene expression | - | IEA | Process |
| GO:0019530 | taurine metabolic process | - | IEA | Process |
| GO:0021510 | spinal cord development | - | IEA | Process |
| GO:0021782 | glial cell development | - | IEA | Process |
| GO:0021915 | neural tube development | - | IEA | Process |
| GO:0022900 | electron transport chain | - | IEA | Process |
| GO:0030060 | L-malate dehydrogenase activity | - | IEA | Function |
| GO:0031175 | neuron projection development | - | IEA | Process |
| GO:0051287 | NAD binding | - | IEA | Function |
| GO:0070062 | extracellular exosome | 19056867.19199708.20458337.23533145. | HDA | Component |
| GO:0070314 | G1 to G0 transition | - | IEA | Process |