
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| LUSC | ||
| ESCA | ||
| KIRP | ||
| PAAD | ||
| BLCA | ||
| GBM | ||
| LIHC | ||
| UVM | ||
| OV |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000266269 | zf-C2H2 | PF00096.26 | 8.2e-26 | 1 | 5 |
| ENSP00000266269 | zf-C2H2 | PF00096.26 | 8.2e-26 | 2 | 5 |
| ENSP00000266269 | zf-C2H2 | PF00096.26 | 8.2e-26 | 3 | 5 |
| ENSP00000266269 | zf-C2H2 | PF00096.26 | 8.2e-26 | 4 | 5 |
| ENSP00000266269 | zf-C2H2 | PF00096.26 | 8.2e-26 | 5 | 5 |
| ENSP00000337520 | zf-C2H2 | PF00096.26 | 1.6e-19 | 1 | 4 |
| ENSP00000337520 | zf-C2H2 | PF00096.26 | 1.6e-19 | 2 | 4 |
| ENSP00000337520 | zf-C2H2 | PF00096.26 | 1.6e-19 | 3 | 4 |
| ENSP00000337520 | zf-C2H2 | PF00096.26 | 1.6e-19 | 4 | 4 |
| ENSP00000384173 | zf-C2H2 | PF00096.26 | 3.9e-16 | 1 | 3 |
| ENSP00000384173 | zf-C2H2 | PF00096.26 | 3.9e-16 | 2 | 3 |
| ENSP00000384173 | zf-C2H2 | PF00096.26 | 3.9e-16 | 3 | 3 |
| ENSP00000215919 | zf-C2H2 | PF00096.26 | 4.8e-15 | 1 | 3 |
| ENSP00000215919 | zf-C2H2 | PF00096.26 | 4.8e-15 | 2 | 3 |
| ENSP00000215919 | zf-C2H2 | PF00096.26 | 4.8e-15 | 3 | 3 |
| ENSP00000266269 | zf-met | PF12874.7 | 7.9e-11 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000494109 | PATZ1-206 | 835 | - | - | - (aa) | - | - |
| ENST00000465287 | PATZ1-205 | 1702 | - | - | - (aa) | - | - |
| ENST00000266269 | PATZ1-202 | 3781 | - | ENSP00000266269 | 687 (aa) | - | Q9HBE1 |
| ENST00000215919 | PATZ1-201 | 2516 | - | ENSP00000215919 | 537 (aa) | - | Q9HBE1 |
| ENST00000405309 | PATZ1-204 | 3711 | - | ENSP00000384173 | 537 (aa) | - | Q9HBE1 |
| ENST00000351933 | PATZ1-203 | 3638 | - | ENSP00000337520 | 641 (aa) | - | Q9HBE1 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000100105 | Platelet Function Tests | 1.5013700E-005 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000100105 | rs572182531 | 22 | 31331345 | CT | Reticulocyte count | 27863252 | [0.024-0.047] unit decrease | 0.03581047 | EFO_0007986 |
| ENSG00000100105 | rs572182531 | 22 | 31331345 | CT | Reticulocyte fraction of red cells | 27863252 | [0.024-0.047] unit decrease | 0.035843 | EFO_0007986 |
| ENSG00000100105 | rs28715 | 22 | 31334183 | T | Red cell distribution width | 28957414 | [0.016-0.031] unit increase | 0.0235931 | EFO_0005192 |
| ENSG00000100105 | rs140074 | 22 | 31329648 | ? | Eosinophil counts | 30595370 | EFO_0004842 |
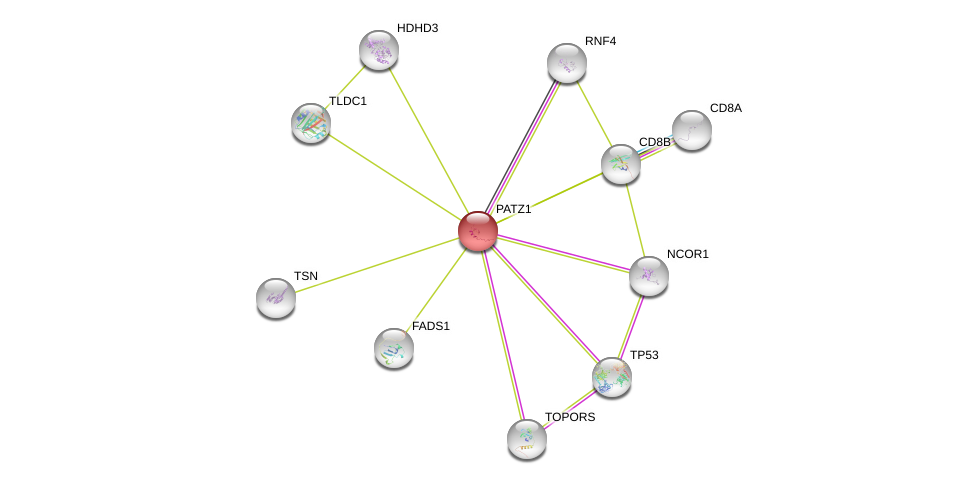
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000100105 | PATZ1 | 99 | 60.038 | ENSAPOG00000018447 | patz1 | 100 | 59.240 | Acanthochromis_polyacanthus |
| ENSG00000100105 | PATZ1 | 100 | 99.127 | ENSAMEG00000007074 | PATZ1 | 100 | 99.127 | Ailuropoda_melanoleuca |
| ENSG00000100105 | PATZ1 | 100 | 60.232 | ENSACIG00000016779 | patz1 | 100 | 59.961 | Amphilophus_citrinellus |
| ENSG00000100105 | PATZ1 | 100 | 57.563 | ENSAPEG00000013354 | patz1 | 100 | 57.703 | Amphiprion_percula |
| ENSG00000100105 | PATZ1 | 100 | 62.549 | ENSATEG00000011260 | patz1 | 100 | 62.353 | Anabas_testudineus |
| ENSG00000100105 | PATZ1 | 85 | 91.267 | ENSAPLG00000003934 | PATZ1 | 99 | 91.096 | Anas_platyrhynchos |
| ENSG00000100105 | PATZ1 | 100 | 100.000 | ENSANAG00000029603 | PATZ1 | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000100105 | PATZ1 | 97 | 62.524 | ENSAMXG00000002273 | patz1 | 98 | 62.147 | Astyanax_mexicanus |
| ENSG00000100105 | PATZ1 | 100 | 99.220 | ENSBTAG00000005478 | PATZ1 | 100 | 99.220 | Bos_taurus |
| ENSG00000100105 | PATZ1 | 100 | 100.000 | ENSCJAG00000010978 | PATZ1 | 100 | 100.000 | Callithrix_jacchus |
| ENSG00000100105 | PATZ1 | 100 | 99.376 | ENSCAFG00000013087 | PATZ1 | 100 | 99.272 | Canis_familiaris |
| ENSG00000100105 | PATZ1 | 100 | 98.752 | ENSCHIG00000020203 | PATZ1 | 100 | 98.752 | Capra_hircus |
| ENSG00000100105 | PATZ1 | 100 | 99.532 | ENSTSYG00000033183 | PATZ1 | 100 | 99.532 | Carlito_syrichta |
| ENSG00000100105 | PATZ1 | 100 | 100.000 | ENSCCAG00000028772 | PATZ1 | 100 | 100.000 | Cebus_capucinus |
| ENSG00000100105 | PATZ1 | 100 | 99.709 | ENSCATG00000006101 | PATZ1 | 100 | 99.709 | Cercocebus_atys |
| ENSG00000100105 | PATZ1 | 100 | 88.986 | ENSCPBG00000011277 | PATZ1 | 100 | 89.130 | Chrysemys_picta_bellii |
| ENSG00000100105 | PATZ1 | 100 | 99.709 | ENSCANG00000024972 | PATZ1 | 100 | 99.709 | Colobus_angolensis_palliatus |
| ENSG00000100105 | PATZ1 | 100 | 97.234 | ENSCGRG00001004071 | Patz1 | 100 | 97.234 | Cricetulus_griseus_chok1gshd |
| ENSG00000100105 | PATZ1 | 93 | 97.978 | ENSCGRG00000004814 | Patz1 | 100 | 97.978 | Cricetulus_griseus_crigri |
| ENSG00000100105 | PATZ1 | 100 | 55.307 | ENSCSEG00000001385 | patz1 | 100 | 55.278 | Cynoglossus_semilaevis |
| ENSG00000100105 | PATZ1 | 100 | 58.997 | ENSCVAG00000007684 | patz1 | 100 | 59.292 | Cyprinodon_variegatus |
| ENSG00000100105 | PATZ1 | 97 | 60.679 | ENSDARG00000076584 | patz1 | 98 | 58.511 | Danio_rerio |
| ENSG00000100105 | PATZ1 | 97 | 95.611 | ENSDNOG00000003183 | PATZ1 | 100 | 98.546 | Dasypus_novemcinctus |
| ENSG00000100105 | PATZ1 | 100 | 99.376 | ENSDORG00000023834 | Patz1 | 100 | 99.376 | Dipodomys_ordii |
| ENSG00000100105 | PATZ1 | 100 | 99.220 | ENSECAG00000025030 | PATZ1 | 100 | 99.220 | Equus_caballus |
| ENSG00000100105 | PATZ1 | 99 | 61.887 | ENSELUG00000002333 | patz1 | 100 | 58.166 | Esox_lucius |
| ENSG00000100105 | PATZ1 | 100 | 99.376 | ENSFCAG00000030253 | PATZ1 | 100 | 99.376 | Felis_catus |
| ENSG00000100105 | PATZ1 | 86 | 91.525 | ENSFALG00000007450 | PATZ1 | 99 | 91.482 | Ficedula_albicollis |
| ENSG00000100105 | PATZ1 | 96 | 98.051 | ENSFDAG00000005614 | PATZ1 | 100 | 98.051 | Fukomys_damarensis |
| ENSG00000100105 | PATZ1 | 100 | 57.918 | ENSFHEG00000014755 | patz1 | 100 | 57.625 | Fundulus_heteroclitus |
| ENSG00000100105 | PATZ1 | 98 | 55.091 | ENSGMOG00000001062 | patz1 | 98 | 54.848 | Gadus_morhua |
| ENSG00000100105 | PATZ1 | 100 | 88.345 | ENSGALG00000006934 | PATZ1 | 100 | 88.489 | Gallus_gallus |
| ENSG00000100105 | PATZ1 | 100 | 59.029 | ENSGAFG00000010053 | patz1 | 100 | 57.856 | Gambusia_affinis |
| ENSG00000100105 | PATZ1 | 85 | 91.938 | ENSGAGG00000003214 | PATZ1 | 100 | 91.767 | Gopherus_agassizii |
| ENSG00000100105 | PATZ1 | 100 | 99.854 | ENSGGOG00000017171 | PATZ1 | 100 | 99.854 | Gorilla_gorilla |
| ENSG00000100105 | PATZ1 | 98 | 62.846 | ENSHCOG00000017644 | patz1 | 98 | 61.372 | Hippocampus_comes |
| ENSG00000100105 | PATZ1 | 97 | 60.285 | ENSIPUG00000017653 | patz1 | 97 | 53.748 | Ictalurus_punctatus |
| ENSG00000100105 | PATZ1 | 100 | 99.127 | ENSSTOG00000020707 | PATZ1 | 100 | 99.127 | Ictidomys_tridecemlineatus |
| ENSG00000100105 | PATZ1 | 100 | 96.807 | ENSJJAG00000015259 | Patz1 | 100 | 96.807 | Jaculus_jaculus |
| ENSG00000100105 | PATZ1 | 100 | 57.670 | ENSKMAG00000008157 | patz1 | 100 | 57.627 | Kryptolebias_marmoratus |
| ENSG00000100105 | PATZ1 | 100 | 98.544 | ENSLAFG00000002317 | PATZ1 | 100 | 98.544 | Loxodonta_africana |
| ENSG00000100105 | PATZ1 | 100 | 99.563 | ENSMFAG00000040968 | PATZ1 | 100 | 99.563 | Macaca_fascicularis |
| ENSG00000100105 | PATZ1 | 100 | 99.563 | ENSMNEG00000037588 | PATZ1 | 100 | 99.563 | Macaca_nemestrina |
| ENSG00000100105 | PATZ1 | 100 | 99.709 | ENSMLEG00000030938 | PATZ1 | 100 | 99.709 | Mandrillus_leucophaeus |
| ENSG00000100105 | PATZ1 | 100 | 57.625 | ENSMAMG00000023652 | patz1 | 100 | 57.771 | Mastacembelus_armatus |
| ENSG00000100105 | PATZ1 | 85 | 87.692 | ENSMGAG00000008439 | PATZ1 | 100 | 87.521 | Meleagris_gallopavo |
| ENSG00000100105 | PATZ1 | 100 | 98.981 | ENSMAUG00000004129 | Patz1 | 100 | 98.981 | Mesocricetus_auratus |
| ENSG00000100105 | PATZ1 | 100 | 99.532 | ENSMICG00000005430 | PATZ1 | 100 | 99.532 | Microcebus_murinus |
| ENSG00000100105 | PATZ1 | 100 | 98.836 | ENSMOCG00000014488 | Patz1 | 100 | 98.836 | Microtus_ochrogaster |
| ENSG00000100105 | PATZ1 | 99 | 55.469 | ENSMMOG00000021679 | patz1 | 100 | 48.656 | Mola_mola |
| ENSG00000100105 | PATZ1 | 100 | 56.941 | ENSMALG00000021172 | patz1 | 100 | 56.657 | Monopterus_albus |
| ENSG00000100105 | PATZ1 | 100 | 98.883 | MGP_CAROLIEiJ_G0016007 | Patz1 | 100 | 98.883 | Mus_caroli |
| ENSG00000100105 | PATZ1 | 100 | 98.981 | ENSMUSG00000020453 | Patz1 | 100 | 98.981 | Mus_musculus |
| ENSG00000100105 | PATZ1 | 100 | 98.981 | MGP_PahariEiJ_G0016884 | Patz1 | 100 | 98.981 | Mus_pahari |
| ENSG00000100105 | PATZ1 | 100 | 98.981 | MGP_SPRETEiJ_G0016846 | Patz1 | 100 | 98.981 | Mus_spretus |
| ENSG00000100105 | PATZ1 | 100 | 99.064 | ENSMLUG00000009942 | PATZ1 | 100 | 99.064 | Myotis_lucifugus |
| ENSG00000100105 | PATZ1 | 100 | 99.272 | ENSNGAG00000010268 | Patz1 | 100 | 99.272 | Nannospalax_galili |
| ENSG00000100105 | PATZ1 | 100 | 60.039 | ENSNBRG00000016032 | patz1 | 100 | 59.768 | Neolamprologus_brichardi |
| ENSG00000100105 | PATZ1 | 68 | 93.978 | ENSMEUG00000016293 | PATZ1 | 99 | 93.966 | Notamacropus_eugenii |
| ENSG00000100105 | PATZ1 | 100 | 98.752 | ENSODEG00000003589 | PATZ1 | 100 | 98.752 | Octodon_degus |
| ENSG00000100105 | PATZ1 | 100 | 58.918 | ENSONIG00000004248 | patz1 | 100 | 58.712 | Oreochromis_niloticus |
| ENSG00000100105 | PATZ1 | 100 | 93.179 | ENSOANG00000014654 | PATZ1 | 100 | 93.469 | Ornithorhynchus_anatinus |
| ENSG00000100105 | PATZ1 | 100 | 58.779 | ENSORLG00020010937 | patz1 | 100 | 58.626 | Oryzias_latipes_hni |
| ENSG00000100105 | PATZ1 | 100 | 58.150 | ENSORLG00015007396 | patz1 | 100 | 58.003 | Oryzias_latipes_hsok |
| ENSG00000100105 | PATZ1 | 100 | 57.459 | ENSOMEG00000006636 | patz1 | 100 | 57.459 | Oryzias_melastigma |
| ENSG00000100105 | PATZ1 | 100 | 99.272 | ENSOGAG00000027905 | PATZ1 | 100 | 99.272 | Otolemur_garnettii |
| ENSG00000100105 | PATZ1 | 100 | 99.854 | ENSPPAG00000033227 | PATZ1 | 100 | 99.854 | Pan_paniscus |
| ENSG00000100105 | PATZ1 | 100 | 99.376 | ENSPPRG00000001573 | PATZ1 | 100 | 99.376 | Panthera_pardus |
| ENSG00000100105 | PATZ1 | 86 | 99.325 | ENSPTIG00000007937 | PATZ1 | 91 | 99.325 | Panthera_tigris_altaica |
| ENSG00000100105 | PATZ1 | 100 | 99.854 | ENSPTRG00000014268 | PATZ1 | 100 | 99.854 | Pan_troglodytes |
| ENSG00000100105 | PATZ1 | 100 | 99.709 | ENSPANG00000006821 | PATZ1 | 100 | 99.709 | Papio_anubis |
| ENSG00000100105 | PATZ1 | 100 | 62.313 | ENSPKIG00000019584 | patz1 | 100 | 61.567 | Paramormyrops_kingsleyae |
| ENSG00000100105 | PATZ1 | 71 | 79.275 | ENSPSIG00000016553 | - | 91 | 82.809 | Pelodiscus_sinensis |
| ENSG00000100105 | PATZ1 | 58 | 95.960 | ENSPSIG00000011562 | - | 100 | 96.571 | Pelodiscus_sinensis |
| ENSG00000100105 | PATZ1 | 100 | 57.022 | ENSPMGG00000010196 | patz1 | 100 | 57.022 | Periophthalmus_magnuspinnatus |
| ENSG00000100105 | PATZ1 | 100 | 98.908 | ENSPEMG00000005321 | Patz1 | 100 | 98.908 | Peromyscus_maniculatus_bairdii |
| ENSG00000100105 | PATZ1 | 100 | 57.478 | ENSPLAG00000010425 | patz1 | 100 | 57.625 | Poecilia_latipinna |
| ENSG00000100105 | PATZ1 | 100 | 57.562 | ENSPMEG00000000255 | patz1 | 100 | 57.709 | Poecilia_mexicana |
| ENSG00000100105 | PATZ1 | 100 | 57.044 | ENSPREG00000002440 | patz1 | 100 | 57.182 | Poecilia_reticulata |
| ENSG00000100105 | PATZ1 | 100 | 99.563 | ENSPPYG00000011732 | PATZ1 | 100 | 99.563 | Pongo_abelii |
| ENSG00000100105 | PATZ1 | 100 | 99.376 | ENSPCOG00000019729 | PATZ1 | 100 | 99.376 | Propithecus_coquereli |
| ENSG00000100105 | PATZ1 | 100 | 99.418 | ENSPVAG00000001749 | PATZ1 | 100 | 99.418 | Pteropus_vampyrus |
| ENSG00000100105 | PATZ1 | 100 | 59.244 | ENSPNYG00000013542 | patz1 | 100 | 58.906 | Pundamilia_nyererei |
| ENSG00000100105 | PATZ1 | 84 | 60.454 | ENSPNAG00000017222 | patz1 | 99 | 59.486 | Pygocentrus_nattereri |
| ENSG00000100105 | PATZ1 | 100 | 99.709 | ENSRBIG00000044231 | PATZ1 | 100 | 99.709 | Rhinopithecus_bieti |
| ENSG00000100105 | PATZ1 | 100 | 99.709 | ENSRROG00000037155 | PATZ1 | 100 | 99.709 | Rhinopithecus_roxellana |
| ENSG00000100105 | PATZ1 | 100 | 100.000 | ENSSBOG00000003894 | PATZ1 | 100 | 100.000 | Saimiri_boliviensis_boliviensis |
| ENSG00000100105 | PATZ1 | 79 | 88.462 | ENSSFOG00015013244 | patz1 | 78 | 88.462 | Scleropages_formosus |
| ENSG00000100105 | PATZ1 | 100 | 87.055 | ENSSPUG00000015298 | PATZ1 | 100 | 86.629 | Sphenodon_punctatus |
| ENSG00000100105 | PATZ1 | 85 | 98.797 | ENSSSCG00000010027 | PATZ1 | 100 | 98.881 | Sus_scrofa |
| ENSG00000100105 | PATZ1 | 100 | 58.264 | ENSTRUG00000015366 | patz1 | 100 | 57.676 | Takifugu_rubripes |
| ENSG00000100105 | PATZ1 | 100 | 99.127 | ENSTTRG00000010279 | PATZ1 | 100 | 99.127 | Tursiops_truncatus |
| ENSG00000100105 | PATZ1 | 100 | 99.376 | ENSUAMG00000015014 | PATZ1 | 100 | 99.376 | Ursus_americanus |
| ENSG00000100105 | PATZ1 | 100 | 73.043 | ENSXETG00000004190 | patz1 | 100 | 72.319 | Xenopus_tropicalis |
| ENSG00000100105 | PATZ1 | 100 | 57.856 | ENSXMAG00000015531 | patz1 | 100 | 58.003 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000978 | RNA polymerase II proximal promoter sequence-specific DNA binding | - | IEA | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | - | ISA | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 19274049. | ISM | Function |
| GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | - | IEA | Function |
| GO:0003677 | DNA binding | 10949935. | NAS | Function |
| GO:0003682 | chromatin binding | - | IEA | Function |
| GO:0005634 | nucleus | 10713105. | TAS | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0006355 | regulation of transcription, DNA-templated | 10713105. | TAS | Process |
| GO:0007283 | spermatogenesis | - | IEA | Process |
| GO:0008584 | male gonad development | - | IEA | Process |
| GO:0030217 | T cell differentiation | - | IEA | Process |
| GO:0045892 | negative regulation of transcription, DNA-templated | 10713105. | TAS | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |