
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 28714518 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| PCPG | |||
| THCA | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| MESO | ||
| ACC | ||
| UCS | ||
| SKCM | ||
| ESCA | ||
| PAAD | ||
| LAML | ||
| LIHC | ||
| LUAD | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000424008 | MICALL1-203 | 918 | - | ENSP00000416766 | 186 (aa) | - | B0QY91 |
| ENST00000445494 | MICALL1-204 | 808 | - | ENSP00000404543 | 210 (aa) | - | B0QY86 |
| ENST00000454685 | MICALL1-205 | 1108 | - | ENSP00000406053 | 370 (aa) | - | H0Y6J8 |
| ENST00000215957 | MICALL1-201 | 4710 | XM_005261791 | ENSP00000215957 | 863 (aa) | XP_005261848 | Q8N3F8 |
| ENST00000489812 | MICALL1-206 | 853 | - | - | - (aa) | - | - |
| ENST00000402631 | MICALL1-202 | 2734 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000100139 | rs34834842 | 22 | 37932590 | G | Platelet distribution width | 27863252 | [0.063-0.099] unit increase | 0.08094431 | EFO_0007984 |
| ENSG00000100139 | rs4254591 | 22 | 37908851 | ? | Heel bone mineral density | 30048462 | [0.013-0.022] unit decrease | 0.0176222 | EFO_0009270 |
| ENSG00000100139 | rs4254591 | 22 | 37908851 | G | Heel bone mineral density | 30598549 | [0.012-0.02] unit decrease | 0.0162358 | EFO_0009270 |
| ENSG00000100139 | rs5750508 | 22 | 37907148 | ? | Body mass index | 30595370 | EFO_0004340 | ||
| ENSG00000100139 | rs3026629 | 22 | 37933278 | A | Smoking status (ever vs never smokers) | 30643258 | [0.0075-0.0158] unit decrease | 0.011653287 | EFO_0004318 |
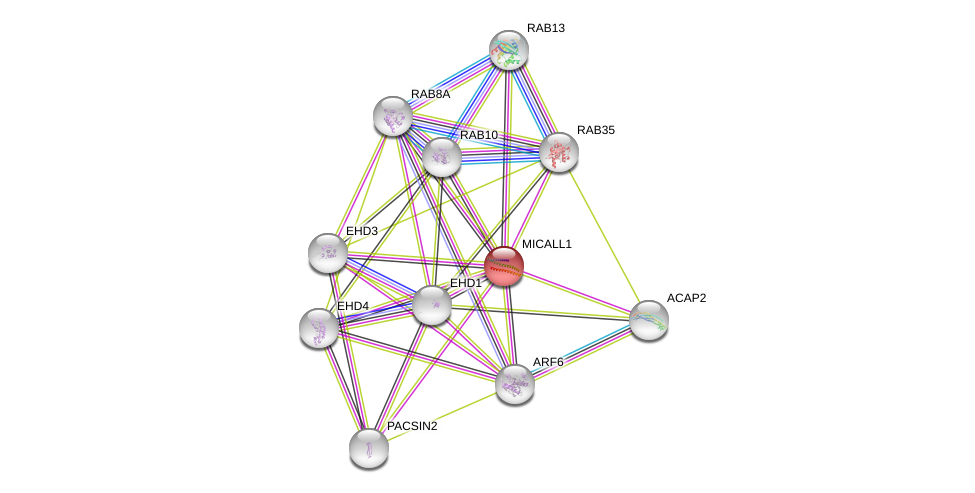
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005515 | protein binding | 19864458.19945174.20801876.21951725.22284051.23596323.27189942. | IPI | Function |
| GO:0005769 | early endosome | 21795389. | IDA | Component |
| GO:0005770 | late endosome | 21795389. | IDA | Component |
| GO:0005802 | trans-Golgi network | 21795389. | IDA | Component |
| GO:0006612 | protein targeting to membrane | 19864458. | IMP | Process |
| GO:0006897 | endocytosis | 20801876. | IMP | Process |
| GO:0006898 | receptor-mediated endocytosis | 21795389. | IMP | Process |
| GO:0017137 | Rab GTPase binding | 19864458.21795389. | IPI | Function |
| GO:0019898 | extrinsic component of membrane | 19864458. | IDA | Component |
| GO:0031175 | neuron projection development | - | ISS | Process |
| GO:0031902 | late endosome membrane | - | IEA | Component |
| GO:0032458 | slow endocytic recycling | 19864458. | IMP | Process |
| GO:0036010 | protein localization to endosome | 23596323. | IMP | Process |
| GO:0042802 | identical protein binding | 23596323. | IPI | Function |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0055038 | recycling endosome membrane | 19864458.21951725. | IDA | Component |
| GO:0055038 | recycling endosome membrane | 21795389. | IDA | Component |
| GO:0070300 | phosphatidic acid binding | 23596323. | IDA | Function |
| GO:0097320 | plasma membrane tubulation | 23596323. | IDA | Process |
| GO:1990090 | cellular response to nerve growth factor stimulus | - | IEA | Process |
| GO:1990126 | retrograde transport, endosome to plasma membrane | 19864458. | IMP | Process |