
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| LGG | |||
| PCPG | |||
| THCA | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| MESO | ||
| ACC | ||
| KIRP | ||
| PAAD | ||
| UCEC | ||
| LIHC | ||
| LUAD | ||
| UVM | ||
| OV |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000391416 | MMR_HSR1 | PF01926.23 | 1.4e-07 | 1 | 1 |
| ENSP00000379703 | MMR_HSR1 | PF01926.23 | 1.4e-07 | 1 | 1 |
| ENSP00000379704 | MMR_HSR1 | PF01926.23 | 1.6e-07 | 1 | 1 |
| ENSP00000494051 | MMR_HSR1 | PF01926.23 | 4.2e-07 | 1 | 1 |
| ENSP00000383956 | MMR_HSR1 | PF01926.23 | 1.6e-05 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000396426 | SEPT3-203 | 2586 | - | ENSP00000379704 | 358 (aa) | - | Q9UH03 |
| ENST00000644076 | SEPT3-207 | 4047 | XM_017028862 | ENSP00000494051 | 856 (aa) | XP_016884351 | A0A2R8Y4H2 |
| ENST00000406029 | SEPT3-204 | 1866 | - | ENSP00000383956 | 294 (aa) | - | B1AHR1 |
| ENST00000460267 | SEPT3-206 | 621 | - | - | - (aa) | - | - |
| ENST00000396425 | SEPT3-202 | 4649 | - | ENSP00000379703 | 350 (aa) | - | Q9UH03 |
| ENST00000449288 | SEPT3-205 | 876 | - | ENSP00000391416 | 178 (aa) | - | B1AHR2 |
| ENST00000396417 | SEPT3-201 | 1896 | - | ENSP00000379695 | 83 (aa) | - | Q9UH03 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa05100 | Bacterial invasion of epithelial cells | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000100167 | rs1062753 | 22 | 41996807 | ? | Response to serotonin reuptake inhibitors in major depressive disorder (plasma drug and metabolite levels) | 24528284 | EFO_0003761|EFO_0005658 | ||
| ENSG00000100167 | rs5751191 | 22 | 41974987 | T | Cognitive performance | 30038396 | [0.016-0.028] unit increase | 0.0221 | EFO_0008354 |
| ENSG00000100167 | rs58874647 | 22 | 41990265 | T | General cognitive ability | 29844566 | z-score increase | 5.934 | EFO_0004337 |
| ENSG00000100167 | rs5751191 | 22 | 41974987 | T | Intelligence | 29942086 | z-score increase | 6.089 | EFO_0004337 |
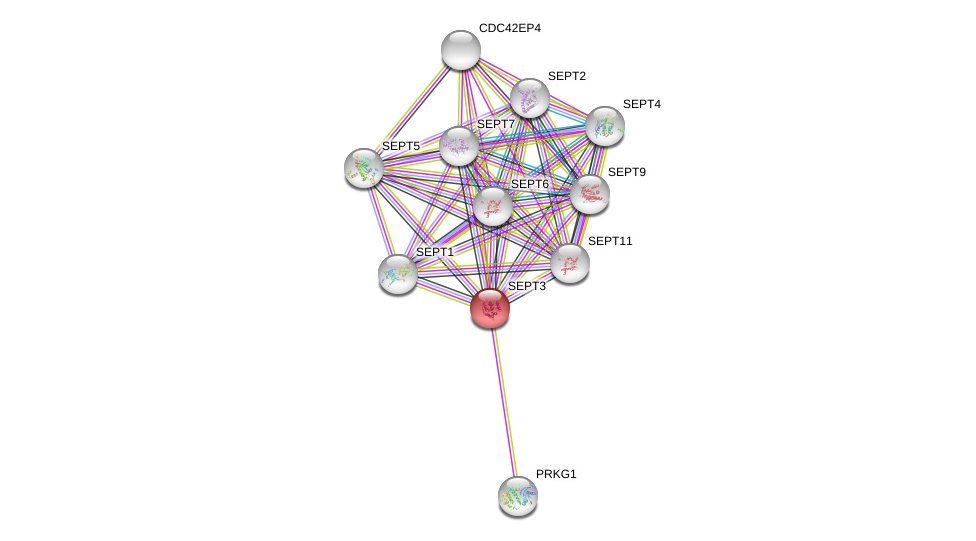
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000100167 | SEPT3 | 93 | 86.447 | ENSAPOG00000003795 | sept3 | 75 | 87.708 | Acanthochromis_polyacanthus |
| ENSG00000100167 | SEPT3 | 100 | 98.980 | ENSAMEG00000009380 | SEPT3 | 99 | 98.873 | Ailuropoda_melanoleuca |
| ENSG00000100167 | SEPT3 | 93 | 84.615 | ENSACIG00000001821 | sept3 | 78 | 85.032 | Amphilophus_citrinellus |
| ENSG00000100167 | SEPT3 | 93 | 85.714 | ENSAOCG00000003633 | sept3 | 75 | 86.218 | Amphiprion_ocellaris |
| ENSG00000100167 | SEPT3 | 93 | 86.081 | ENSAPEG00000022582 | sept3 | 99 | 84.698 | Amphiprion_percula |
| ENSG00000100167 | SEPT3 | 93 | 84.615 | ENSATEG00000000191 | sept3 | 96 | 78.448 | Anabas_testudineus |
| ENSG00000100167 | SEPT3 | 100 | 97.191 | ENSAPLG00000002801 | SEPT3 | 99 | 96.108 | Anas_platyrhynchos |
| ENSG00000100167 | SEPT3 | 100 | 96.067 | ENSACAG00000003974 | SEPT3 | 99 | 94.556 | Anolis_carolinensis |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSANAG00000019445 | SEPT3 | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000100167 | SEPT3 | 93 | 85.348 | ENSACLG00000009596 | - | 83 | 82.051 | Astatotilapia_calliptera |
| ENSG00000100167 | SEPT3 | 100 | 85.714 | ENSAMXG00000018729 | sept3 | 99 | 84.661 | Astyanax_mexicanus |
| ENSG00000100167 | SEPT3 | 100 | 98.980 | ENSBTAG00000013173 | SEPT3 | 100 | 98.883 | Bos_taurus |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSCJAG00000043636 | SEPT3 | 100 | 99.660 | Callithrix_jacchus |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSCAFG00000000995 | SEPT3 | 99 | 98.623 | Canis_familiaris |
| ENSG00000100167 | SEPT3 | 96 | 86.550 | ENSCAFG00020000881 | - | 99 | 77.835 | Canis_lupus_dingo |
| ENSG00000100167 | SEPT3 | 100 | 98.980 | ENSCHIG00000016257 | SEPT3 | 100 | 98.883 | Capra_hircus |
| ENSG00000100167 | SEPT3 | 100 | 99.660 | ENSTSYG00000012514 | SEPT3 | 100 | 99.660 | Carlito_syrichta |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSCAPG00000017776 | - | 100 | 98.333 | Cavia_aperea |
| ENSG00000100167 | SEPT3 | 100 | 87.989 | ENSCAPG00000007508 | - | 100 | 87.989 | Cavia_aperea |
| ENSG00000100167 | SEPT3 | 99 | 85.593 | ENSCPOG00000004011 | - | 99 | 85.593 | Cavia_porcellus |
| ENSG00000100167 | SEPT3 | 100 | 99.441 | ENSCPOG00000013212 | - | 100 | 99.441 | Cavia_porcellus |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSCCAG00000022439 | SEPT3 | 100 | 100.000 | Cebus_capucinus |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSCATG00000045173 | SEPT3 | 100 | 100.000 | Cercocebus_atys |
| ENSG00000100167 | SEPT3 | 100 | 98.639 | ENSCLAG00000017190 | SEPT3 | 100 | 99.110 | Chinchilla_lanigera |
| ENSG00000100167 | SEPT3 | 100 | 99.721 | ENSCSAG00000005722 | SEPT3 | 100 | 99.721 | Chlorocebus_sabaeus |
| ENSG00000100167 | SEPT3 | 100 | 97.191 | ENSCPBG00000020516 | SEPT3 | 99 | 93.983 | Chrysemys_picta_bellii |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSCANG00000030048 | SEPT3 | 100 | 99.718 | Colobus_angolensis_palliatus |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSCGRG00001018092 | Sept3 | 100 | 99.162 | Cricetulus_griseus_chok1gshd |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSCGRG00000016977 | Sept3 | 100 | 99.703 | Cricetulus_griseus_crigri |
| ENSG00000100167 | SEPT3 | 93 | 85.662 | ENSCVAG00000023430 | sept3 | 76 | 85.669 | Cyprinodon_variegatus |
| ENSG00000100167 | SEPT3 | 93 | 86.813 | ENSDARG00000030656 | sept3 | 99 | 78.670 | Danio_rerio |
| ENSG00000100167 | SEPT3 | 100 | 97.191 | ENSDNOG00000032372 | SEPT3 | 97 | 92.973 | Dasypus_novemcinctus |
| ENSG00000100167 | SEPT3 | 96 | 98.246 | ENSDORG00000014056 | Sept3 | 100 | 87.465 | Dipodomys_ordii |
| ENSG00000100167 | SEPT3 | 80 | 97.203 | ENSETEG00000004487 | SEPT3 | 79 | 100.000 | Echinops_telfairi |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSEASG00005001015 | - | 99 | 99.713 | Equus_asinus_asinus |
| ENSG00000100167 | SEPT3 | 100 | 99.714 | ENSECAG00000023601 | SEPT3 | 100 | 99.714 | Equus_caballus |
| ENSG00000100167 | SEPT3 | 100 | 98.315 | ENSEEUG00000011889 | SEPT3 | 100 | 99.074 | Erinaceus_europaeus |
| ENSG00000100167 | SEPT3 | 93 | 87.546 | ENSELUG00000003004 | sept3 | 99 | 76.471 | Esox_lucius |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSFCAG00000000889 | SEPT3 | 100 | 100.000 | Felis_catus |
| ENSG00000100167 | SEPT3 | 100 | 96.067 | ENSFALG00000011773 | SEPT3 | 99 | 94.269 | Ficedula_albicollis |
| ENSG00000100167 | SEPT3 | 100 | 98.876 | ENSFDAG00000010054 | SEPT3 | 100 | 99.110 | Fukomys_damarensis |
| ENSG00000100167 | SEPT3 | 93 | 84.982 | ENSFHEG00000005258 | sept3 | 77 | 84.424 | Fundulus_heteroclitus |
| ENSG00000100167 | SEPT3 | 93 | 83.516 | ENSGMOG00000005438 | sept3 | 99 | 76.389 | Gadus_morhua |
| ENSG00000100167 | SEPT3 | 100 | 95.506 | ENSGALG00000011913 | SEPT3 | 99 | 93.123 | Gallus_gallus |
| ENSG00000100167 | SEPT3 | 93 | 86.081 | ENSGAFG00000008333 | sept3 | 75 | 86.538 | Gambusia_affinis |
| ENSG00000100167 | SEPT3 | 97 | 84.393 | ENSGACG00000015988 | sept3 | 99 | 80.851 | Gasterosteus_aculeatus |
| ENSG00000100167 | SEPT3 | 92 | 98.148 | ENSGAGG00000005342 | - | 98 | 95.105 | Gopherus_agassizii |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSGGOG00000008961 | SEPT3 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000100167 | SEPT3 | 93 | 84.982 | ENSHBUG00000005002 | sept3 | 75 | 85.350 | Haplochromis_burtoni |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSHGLG00000006998 | - | 100 | 99.703 | Heterocephalus_glaber_female |
| ENSG00000100167 | SEPT3 | 100 | 90.449 | ENSHGLG00000020767 | - | 90 | 75.676 | Heterocephalus_glaber_female |
| ENSG00000100167 | SEPT3 | 100 | 88.764 | ENSHGLG00100019917 | - | 90 | 74.775 | Heterocephalus_glaber_male |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSHGLG00100007916 | - | 100 | 99.703 | Heterocephalus_glaber_male |
| ENSG00000100167 | SEPT3 | 92 | 74.539 | ENSHCOG00000016310 | sept3 | 76 | 73.929 | Hippocampus_comes |
| ENSG00000100167 | SEPT3 | 100 | 83.607 | ENSIPUG00000021317 | sept3 | 99 | 83.529 | Ictalurus_punctatus |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSSTOG00000012963 | SEPT3 | 100 | 99.437 | Ictidomys_tridecemlineatus |
| ENSG00000100167 | SEPT3 | 100 | 97.279 | ENSJJAG00000019615 | Sept3 | 99 | 97.458 | Jaculus_jaculus |
| ENSG00000100167 | SEPT3 | 93 | 84.982 | ENSKMAG00000021717 | sept3 | 76 | 85.577 | Kryptolebias_marmoratus |
| ENSG00000100167 | SEPT3 | 93 | 84.249 | ENSLBEG00000019948 | sept3 | 74 | 84.713 | Labrus_bergylta |
| ENSG00000100167 | SEPT3 | 96 | 75.897 | ENSLOCG00000011996 | sept3 | 100 | 77.436 | Lepisosteus_oculatus |
| ENSG00000100167 | SEPT3 | 100 | 97.973 | ENSLAFG00000015724 | SEPT3 | 99 | 98.319 | Loxodonta_africana |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSMFAG00000039306 | SEPT3 | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSMMUG00000015139 | SEPT3 | 100 | 100.000 | Macaca_mulatta |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSMNEG00000038786 | SEPT3 | 100 | 100.000 | Macaca_nemestrina |
| ENSG00000100167 | SEPT3 | 92 | 85.185 | ENSMAMG00000004114 | sept3 | 73 | 86.424 | Mastacembelus_armatus |
| ENSG00000100167 | SEPT3 | 93 | 85.348 | ENSMZEG00005023846 | sept3 | 76 | 85.669 | Maylandia_zebra |
| ENSG00000100167 | SEPT3 | 100 | 95.506 | ENSMGAG00000012105 | SEPT3 | 89 | 95.224 | Meleagris_gallopavo |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSMAUG00000014933 | Sept3 | 100 | 99.703 | Mesocricetus_auratus |
| ENSG00000100167 | SEPT3 | 100 | 99.660 | ENSMICG00000003589 | SEPT3 | 100 | 99.660 | Microcebus_murinus |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSMOCG00000015408 | Sept3 | 100 | 98.873 | Microtus_ochrogaster |
| ENSG00000100167 | SEPT3 | 100 | 83.889 | ENSMMOG00000008339 | sept3 | 99 | 83.333 | Mola_mola |
| ENSG00000100167 | SEPT3 | 93 | 84.672 | ENSMALG00000000364 | sept3 | 98 | 83.871 | Monopterus_albus |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | MGP_CAROLIEiJ_G0020078 | Sept3 | 100 | 99.703 | Mus_caroli |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSMUSG00000022456 | Sept3 | 100 | 99.703 | Mus_musculus |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | MGP_PahariEiJ_G0020080 | Sept3 | 100 | 99.703 | Mus_pahari |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | MGP_SPRETEiJ_G0020978 | Sept3 | 100 | 99.703 | Mus_spretus |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSMPUG00000016339 | SEPT3 | 99 | 85.714 | Mustela_putorius_furo |
| ENSG00000100167 | SEPT3 | 100 | 98.876 | ENSMLUG00000002571 | SEPT3 | 99 | 98.310 | Myotis_lucifugus |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSNGAG00000020671 | Sept3 | 100 | 99.407 | Nannospalax_galili |
| ENSG00000100167 | SEPT3 | 92 | 86.585 | ENSNBRG00000018345 | sept3 | 98 | 84.375 | Neolamprologus_brichardi |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSNLEG00000015691 | SEPT3 | 100 | 99.320 | Nomascus_leucogenys |
| ENSG00000100167 | SEPT3 | 100 | 98.876 | ENSOPRG00000008638 | SEPT3 | 100 | 98.324 | Ochotona_princeps |
| ENSG00000100167 | SEPT3 | 100 | 98.876 | ENSODEG00000012406 | SEPT3 | 100 | 99.143 | Octodon_degus |
| ENSG00000100167 | SEPT3 | 100 | 80.328 | ENSONIG00000018012 | sept3 | 99 | 81.948 | Oreochromis_niloticus |
| ENSG00000100167 | SEPT3 | 100 | 95.000 | ENSOANG00000004244 | SEPT3 | 98 | 94.946 | Ornithorhynchus_anatinus |
| ENSG00000100167 | SEPT3 | 100 | 98.876 | ENSOCUG00000008260 | SEPT3 | 100 | 78.680 | Oryctolagus_cuniculus |
| ENSG00000100167 | SEPT3 | 93 | 84.982 | ENSORLG00000004172 | sept3 | 91 | 75.723 | Oryzias_latipes |
| ENSG00000100167 | SEPT3 | 93 | 84.249 | ENSORLG00020009259 | sept3 | 79 | 84.295 | Oryzias_latipes_hni |
| ENSG00000100167 | SEPT3 | 93 | 84.982 | ENSORLG00015007295 | sept3 | 99 | 79.240 | Oryzias_latipes_hsok |
| ENSG00000100167 | SEPT3 | 93 | 85.348 | ENSOMEG00000000404 | sept3 | 97 | 84.532 | Oryzias_melastigma |
| ENSG00000100167 | SEPT3 | 100 | 99.441 | ENSOGAG00000014752 | SEPT3 | 100 | 99.441 | Otolemur_garnettii |
| ENSG00000100167 | SEPT3 | 100 | 99.320 | ENSOARG00000018800 | SEPT3 | 100 | 98.883 | Ovis_aries |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSPPAG00000013116 | SEPT3 | 100 | 100.000 | Pan_paniscus |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSPPRG00000002117 | SEPT3 | 100 | 100.000 | Panthera_pardus |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSPTIG00000011548 | SEPT3 | 99 | 100.000 | Panthera_tigris_altaica |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSPTRG00000014444 | SEPT3 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSPANG00000009994 | - | 100 | 100.000 | Papio_anubis |
| ENSG00000100167 | SEPT3 | 100 | 79.695 | ENSPKIG00000002056 | sept3 | 99 | 80.337 | Paramormyrops_kingsleyae |
| ENSG00000100167 | SEPT3 | 100 | 96.629 | ENSPSIG00000012269 | SEPT3 | 99 | 95.536 | Pelodiscus_sinensis |
| ENSG00000100167 | SEPT3 | 96 | 81.871 | ENSPMGG00000006200 | sept3 | 78 | 82.209 | Periophthalmus_magnuspinnatus |
| ENSG00000100167 | SEPT3 | 100 | 98.980 | ENSPEMG00000009916 | Sept3 | 100 | 99.407 | Peromyscus_maniculatus_bairdii |
| ENSG00000100167 | SEPT3 | 100 | 96.067 | ENSPCIG00000004039 | SEPT3 | 99 | 94.048 | Phascolarctos_cinereus |
| ENSG00000100167 | SEPT3 | 100 | 84.916 | ENSPFOG00000014541 | sept3 | 99 | 81.356 | Poecilia_formosa |
| ENSG00000100167 | SEPT3 | 93 | 86.081 | ENSPLAG00000010228 | sept3 | 74 | 86.538 | Poecilia_latipinna |
| ENSG00000100167 | SEPT3 | 93 | 86.081 | ENSPMEG00000019416 | sept3 | 73 | 86.538 | Poecilia_mexicana |
| ENSG00000100167 | SEPT3 | 93 | 85.714 | ENSPREG00000012774 | sept3 | 95 | 86.218 | Poecilia_reticulata |
| ENSG00000100167 | SEPT3 | 100 | 100.000 | ENSPPYG00000011886 | SEPT3 | 94 | 100.000 | Pongo_abelii |
| ENSG00000100167 | SEPT3 | 100 | 99.660 | ENSPCOG00000024794 | SEPT3 | 100 | 99.660 | Propithecus_coquereli |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSPVAG00000001191 | SEPT3 | 100 | 98.603 | Pteropus_vampyrus |
| ENSG00000100167 | SEPT3 | 93 | 84.982 | ENSPNYG00000023957 | sept3 | 77 | 85.350 | Pundamilia_nyererei |
| ENSG00000100167 | SEPT3 | 100 | 86.111 | ENSPNAG00000023001 | sept3 | 99 | 82.596 | Pygocentrus_nattereri |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSRNOG00000007686 | Sept3 | 100 | 98.324 | Rattus_norvegicus |
| ENSG00000100167 | SEPT3 | 100 | 99.660 | ENSRBIG00000040851 | SEPT3 | 100 | 99.660 | Rhinopithecus_bieti |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSRROG00000042463 | SEPT3 | 100 | 92.157 | Rhinopithecus_roxellana |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSSBOG00000024825 | SEPT3 | 99 | 96.092 | Saimiri_boliviensis_boliviensis |
| ENSG00000100167 | SEPT3 | 100 | 96.067 | ENSSHAG00000012446 | SEPT3 | 97 | 90.986 | Sarcophilus_harrisii |
| ENSG00000100167 | SEPT3 | 93 | 88.278 | ENSSFOG00015011047 | sept3 | 99 | 80.165 | Scleropages_formosus |
| ENSG00000100167 | SEPT3 | 93 | 84.249 | ENSSMAG00000013826 | sept3 | 76 | 84.615 | Scophthalmus_maximus |
| ENSG00000100167 | SEPT3 | 93 | 86.081 | ENSSDUG00000004549 | sept3 | 75 | 86.306 | Seriola_dumerili |
| ENSG00000100167 | SEPT3 | 93 | 86.081 | ENSSLDG00000023869 | sept3 | 75 | 86.306 | Seriola_lalandi_dorsalis |
| ENSG00000100167 | SEPT3 | 100 | 94.944 | ENSSPUG00000005948 | SEPT3 | 95 | 94.412 | Sphenodon_punctatus |
| ENSG00000100167 | SEPT3 | 100 | 98.980 | ENSSSCG00000000040 | SEPT3 | 100 | 98.603 | Sus_scrofa |
| ENSG00000100167 | SEPT3 | 91 | 97.753 | ENSTGUG00000009729 | SEPT3 | 100 | 98.630 | Taeniopygia_guttata |
| ENSG00000100167 | SEPT3 | 93 | 85.348 | ENSTRUG00000000028 | sept3 | 99 | 80.896 | Takifugu_rubripes |
| ENSG00000100167 | SEPT3 | 93 | 84.982 | ENSTNIG00000004228 | sept3 | 99 | 84.395 | Tetraodon_nigroviridis |
| ENSG00000100167 | SEPT3 | 100 | 99.162 | ENSTTRG00000017358 | SEPT3 | 100 | 99.162 | Tursiops_truncatus |
| ENSG00000100167 | SEPT3 | 83 | 82.993 | ENSUAMG00000003202 | - | 55 | 95.161 | Ursus_americanus |
| ENSG00000100167 | SEPT3 | 100 | 99.438 | ENSUMAG00000020618 | - | 100 | 99.162 | Ursus_maritimus |
| ENSG00000100167 | SEPT3 | 100 | 99.441 | ENSVVUG00000002589 | - | 100 | 99.441 | Vulpes_vulpes |
| ENSG00000100167 | SEPT3 | 100 | 91.573 | ENSXETG00000000261 | sept3 | 99 | 90.751 | Xenopus_tropicalis |
| ENSG00000100167 | SEPT3 | 93 | 85.714 | ENSXCOG00000013650 | sept3 | 75 | 86.218 | Xiphophorus_couchianus |
| ENSG00000100167 | SEPT3 | 93 | 85.714 | ENSXMAG00000017559 | sept3 | 94 | 85.714 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003674 | molecular_function | - | ND | Function |
| GO:0003924 | GTPase activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 25416956. | IPI | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005940 | septin ring | 21873635. | IBA | Component |
| GO:0015630 | microtubule cytoskeleton | 21873635. | IBA | Component |
| GO:0030054 | cell junction | - | IEA | Component |
| GO:0031105 | septin complex | 21873635. | IBA | Component |
| GO:0031105 | septin complex | - | ISS | Component |
| GO:0042802 | identical protein binding | 25416956. | IPI | Function |
| GO:0043005 | neuron projection | 21873635. | IBA | Component |
| GO:0060090 | molecular adaptor activity | 21873635. | IBA | Function |
| GO:0061640 | cytoskeleton-dependent cytokinesis | 21873635. | IBA | Process |
| GO:0098793 | presynapse | - | ISS | Component |
| GO:0099569 | presynaptic cytoskeleton | 21873635. | IBA | Component |