
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THYM | |||||||
| THYM | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| PCPG | |||
| THCA | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| ACC | ||
| UCS | ||
| HNSC | ||
| SKCM | ||
| ESCA | ||
| BLCA | ||
| CESC | ||
| READ | ||
| GBM | ||
| LIHC | ||
| LGG | ||
| LUAD |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000491244 | DEAD | PF00270.29 | 4.8e-49 | 1 | 1 |
| ENSP00000385536 | DEAD | PF00270.29 | 4.8e-49 | 1 | 1 |
| ENSP00000491776 | DEAD | PF00270.29 | 4.8e-49 | 1 | 1 |
| ENSP00000380033 | DEAD | PF00270.29 | 4.8e-49 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000467279 | DDX17-206 | 712 | - | - | - (aa) | - | - |
| ENST00000475004 | DDX17-207 | 443 | - | ENSP00000489029 | 56 (aa) | - | A0A0U1RQJ0 |
| ENST00000431312 | DDX17-204 | 1853 | - | - | - (aa) | - | - |
| ENST00000640332 | DDX17-211 | 2190 | - | ENSP00000491244 | 729 (aa) | - | Q92841 |
| ENST00000432525 | DDX17-205 | 1354 | - | - | - (aa) | - | - |
| ENST00000477112 | DDX17-208 | 654 | - | - | - (aa) | - | - |
| ENST00000216019 | DDX17-201 | 6441 | - | - | - (aa) | - | - |
| ENST00000479734 | DDX17-209 | 441 | - | - | - (aa) | - | - |
| ENST00000396821 | DDX17-202 | 4791 | - | ENSP00000380033 | 731 (aa) | - | UPI0001AE634C |
| ENST00000403230 | DDX17-203 | 2322 | - | ENSP00000385536 | 729 (aa) | - | A0A1X7SBZ2 |
| ENST00000640668 | DDX17-212 | 2196 | - | ENSP00000491776 | 731 (aa) | - | A0A1W2PQ51 |
| ENST00000497196 | DDX17-210 | 856 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000100201 | Child Development Disorders, Pervasive | 3.1330000E-005 | - |
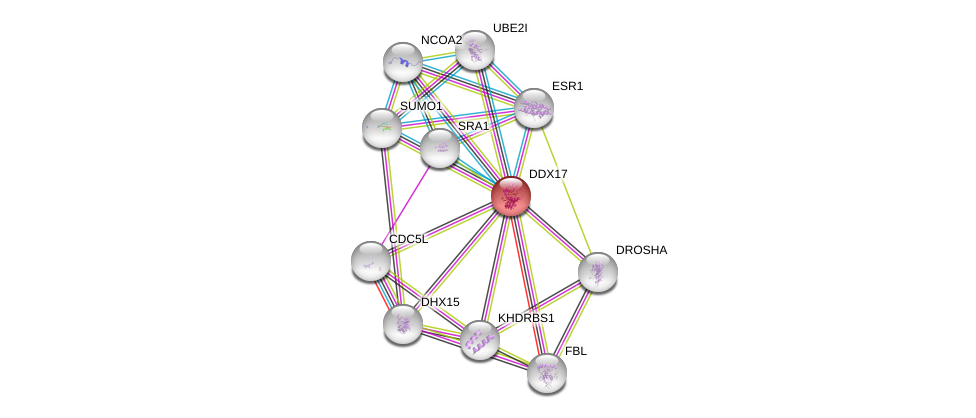
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000100201 | DDX17 | 52 | 30.909 | ENSG00000125485 | DDX31 | 61 | 30.110 |
| ENSG00000100201 | DDX17 | 73 | 51.220 | ENSG00000213782 | DDX47 | 71 | 41.194 |
| ENSG00000100201 | DDX17 | 60 | 43.333 | ENSG00000184735 | DDX53 | 70 | 43.596 |
| ENSG00000100201 | DDX17 | 63 | 57.143 | ENSG00000080007 | DDX43 | 68 | 42.637 |
| ENSG00000100201 | DDX17 | 71 | 90.000 | ENSG00000108654 | DDX5 | 100 | 89.759 |
| ENSG00000100201 | DDX17 | 50 | 32.337 | ENSG00000123064 | DDX54 | 99 | 35.577 |
| ENSG00000100201 | DDX17 | 50 | 30.053 | ENSG00000157349 | DDX19B | 96 | 30.053 |
| ENSG00000100201 | DDX17 | 68 | 52.632 | ENSG00000141543 | EIF4A3 | 82 | 35.277 |
| ENSG00000100201 | DDX17 | 50 | 33.784 | ENSG00000124228 | DDX27 | 88 | 31.429 |
| ENSG00000100201 | DDX17 | 53 | 39.568 | ENSG00000174243 | DDX23 | 55 | 40.000 |
| ENSG00000100201 | DDX17 | 50 | 34.218 | ENSG00000165732 | DDX21 | 51 | 33.952 |
| ENSG00000100201 | DDX17 | 63 | 65.714 | ENSG00000215301 | DDX3X | 98 | 41.260 |
| ENSG00000100201 | DDX17 | 57 | 34.450 | ENSG00000118197 | DDX59 | 98 | 32.127 |
| ENSG00000100201 | DDX17 | 75 | 45.238 | ENSG00000178105 | DDX10 | 56 | 32.987 |
| ENSG00000100201 | DDX17 | 77 | 53.488 | ENSG00000183258 | DDX41 | 65 | 39.952 |
| ENSG00000100201 | DDX17 | 71 | 47.500 | ENSG00000198231 | DDX42 | 61 | 43.880 |
| ENSG00000100201 | DDX17 | 63 | 51.429 | ENSG00000156976 | EIF4A2 | 90 | 33.832 |
| ENSG00000100201 | DDX17 | 55 | 30.340 | ENSG00000278053 | DDX52 | 67 | 30.216 |
| ENSG00000100201 | DDX17 | 63 | 51.429 | ENSG00000161960 | EIF4A1 | 97 | 34.188 |
| ENSG00000100201 | DDX17 | 71 | 57.500 | ENSG00000067048 | DDX3Y | 71 | 39.072 |
| ENSG00000100201 | DDX17 | 73 | 48.780 | ENSG00000107625 | DDX50 | 50 | 34.417 |
| ENSG00000100201 | DDX17 | 50 | 32.454 | ENSG00000109832 | DDX25 | 93 | 35.000 |
| ENSG00000100201 | DDX17 | 52 | 30.971 | ENSG00000110367 | DDX6 | 77 | 30.971 |
| ENSG00000100201 | DDX17 | 63 | 60.000 | ENSG00000152670 | DDX4 | 68 | 39.798 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSAPOG00000017303 | si:dkey-156n14.5 | 93 | 74.641 | Acanthochromis_polyacanthus |
| ENSG00000100201 | DDX17 | 100 | 99.453 | ENSAMEG00000010601 | DDX17 | 100 | 99.453 | Ailuropoda_melanoleuca |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSAOCG00000004874 | si:dkey-156n14.5 | 93 | 74.290 | Amphiprion_ocellaris |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSAPEG00000018317 | si:dkey-156n14.5 | 94 | 74.062 | Amphiprion_percula |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSATEG00000021733 | si:dkey-156n14.5 | 95 | 73.892 | Anabas_testudineus |
| ENSG00000100201 | DDX17 | 71 | 100.000 | ENSAPLG00000014334 | - | 99 | 88.552 | Anas_platyrhynchos |
| ENSG00000100201 | DDX17 | 100 | 91.530 | ENSANAG00000035052 | DDX17 | 100 | 92.496 | Aotus_nancymaae |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSACLG00000015882 | DDX17 | 94 | 75.654 | Astatotilapia_calliptera |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSACLG00000015887 | si:dkey-156n14.5 | 93 | 73.865 | Astatotilapia_calliptera |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSBTAG00000009887 | DDX17 | 100 | 99.692 | Bos_taurus |
| ENSG00000100201 | DDX17 | 71 | 72.500 | WBGene00010260 | ddx-17 | 79 | 60.497 | Caenorhabditis_elegans |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSCJAG00000011797 | DDX17 | 100 | 99.846 | Callithrix_jacchus |
| ENSG00000100201 | DDX17 | 97 | 99.295 | ENSCAFG00000001395 | DDX17 | 100 | 99.846 | Canis_familiaris |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSCAFG00020002867 | DDX17 | 100 | 99.846 | Canis_lupus_dingo |
| ENSG00000100201 | DDX17 | 100 | 99.314 | ENSCHIG00000021332 | DDX17 | 100 | 99.314 | Capra_hircus |
| ENSG00000100201 | DDX17 | 100 | 98.769 | ENSTSYG00000001303 | DDX17 | 99 | 98.769 | Carlito_syrichta |
| ENSG00000100201 | DDX17 | 100 | 99.314 | ENSCPOG00000011790 | DDX17 | 100 | 99.314 | Cavia_porcellus |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSCCAG00000024116 | DDX17 | 100 | 100.000 | Cebus_capucinus |
| ENSG00000100201 | DDX17 | 100 | 100.000 | ENSCATG00000035701 | DDX17 | 99 | 100.000 | Cercocebus_atys |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSCLAG00000017294 | DDX17 | 100 | 99.540 | Chinchilla_lanigera |
| ENSG00000100201 | DDX17 | 89 | 100.000 | ENSCSAG00000006649 | DDX17 | 100 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000100201 | DDX17 | 100 | 97.942 | ENSCHOG00000001931 | DDX17 | 100 | 97.942 | Choloepus_hoffmanni |
| ENSG00000100201 | DDX17 | 63 | 71.429 | ENSCING00000008750 | - | 82 | 45.781 | Ciona_intestinalis |
| ENSG00000100201 | DDX17 | 71 | 92.500 | ENSCING00000004147 | - | 90 | 62.715 | Ciona_intestinalis |
| ENSG00000100201 | DDX17 | 100 | 99.863 | ENSCANG00000002348 | DDX17 | 100 | 99.863 | Colobus_angolensis_palliatus |
| ENSG00000100201 | DDX17 | 89 | 99.080 | ENSCGRG00001023050 | Ddx17 | 100 | 99.080 | Cricetulus_griseus_chok1gshd |
| ENSG00000100201 | DDX17 | 89 | 99.080 | ENSCGRG00000007756 | Ddx17 | 100 | 99.080 | Cricetulus_griseus_crigri |
| ENSG00000100201 | DDX17 | 75 | 95.238 | ENSCSEG00000011351 | si:dkey-156n14.5 | 91 | 89.125 | Cynoglossus_semilaevis |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSCVAG00000010921 | si:dkey-156n14.5 | 95 | 73.035 | Cyprinodon_variegatus |
| ENSG00000100201 | DDX17 | 73 | 97.561 | ENSDARG00000010873 | DDX17 | 91 | 89.125 | Danio_rerio |
| ENSG00000100201 | DDX17 | 99 | 98.755 | ENSDNOG00000006499 | DDX17 | 99 | 98.755 | Dasypus_novemcinctus |
| ENSG00000100201 | DDX17 | 100 | 96.296 | ENSDORG00000003358 | Ddx17 | 100 | 99.233 | Dipodomys_ordii |
| ENSG00000100201 | DDX17 | 75 | 66.667 | FBgn0035720 | CG10077 | 78 | 65.768 | Drosophila_melanogaster |
| ENSG00000100201 | DDX17 | 56 | 95.333 | ENSETEG00000009484 | - | 63 | 95.070 | Echinops_telfairi |
| ENSG00000100201 | DDX17 | 89 | 99.538 | ENSEASG00005018767 | DDX17 | 100 | 99.538 | Equus_asinus_asinus |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSECAG00000025110 | DDX17 | 100 | 99.538 | Equus_caballus |
| ENSG00000100201 | DDX17 | 100 | 99.316 | ENSFCAG00000031570 | DDX17 | 96 | 100.000 | Felis_catus |
| ENSG00000100201 | DDX17 | 85 | 88.854 | ENSFALG00000012560 | DDX17 | 84 | 88.264 | Ficedula_albicollis |
| ENSG00000100201 | DDX17 | 87 | 99.370 | ENSFDAG00000009967 | DDX17 | 100 | 99.370 | Fukomys_damarensis |
| ENSG00000100201 | DDX17 | 87 | 89.252 | ENSGALG00000037360 | DDX17 | 95 | 88.462 | Gallus_gallus |
| ENSG00000100201 | DDX17 | 71 | 95.000 | ENSGACG00000019151 | si:dkey-156n14.5 | 85 | 69.759 | Gasterosteus_aculeatus |
| ENSG00000100201 | DDX17 | 100 | 100.000 | ENSGGOG00000008309 | DDX17 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSHBUG00000021303 | si:dkey-156n14.5 | 93 | 73.709 | Haplochromis_burtoni |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSHBUG00000022268 | DDX17 | 94 | 75.367 | Haplochromis_burtoni |
| ENSG00000100201 | DDX17 | 100 | 98.903 | ENSHGLG00000012568 | DDX17 | 100 | 98.903 | Heterocephalus_glaber_female |
| ENSG00000100201 | DDX17 | 100 | 98.903 | ENSHGLG00100011291 | DDX17 | 100 | 98.903 | Heterocephalus_glaber_male |
| ENSG00000100201 | DDX17 | 73 | 97.561 | ENSIPUG00000015768 | si:dkey-156n14.5 | 94 | 89.920 | Ictalurus_punctatus |
| ENSG00000100201 | DDX17 | 100 | 99.177 | ENSSTOG00000013699 | DDX17 | 100 | 99.177 | Ictidomys_tridecemlineatus |
| ENSG00000100201 | DDX17 | 99 | 98.483 | ENSJJAG00000011350 | Ddx17 | 100 | 98.483 | Jaculus_jaculus |
| ENSG00000100201 | DDX17 | 100 | 99.863 | ENSMFAG00000046051 | DDX17 | 99 | 100.000 | Macaca_fascicularis |
| ENSG00000100201 | DDX17 | 100 | 99.863 | ENSMMUG00000004304 | DDX17 | 100 | 99.863 | Macaca_mulatta |
| ENSG00000100201 | DDX17 | 100 | 99.863 | ENSMNEG00000022218 | DDX17 | 99 | 100.000 | Macaca_nemestrina |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSMLEG00000029141 | DDX17 | 99 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSMAMG00000008427 | si:dkey-156n14.5 | 93 | 73.034 | Mastacembelus_armatus |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSMZEG00005009937 | si:dkey-156n14.5 | 93 | 73.865 | Maylandia_zebra |
| ENSG00000100201 | DDX17 | 87 | 89.286 | ENSMGAG00000012423 | DDX17 | 86 | 98.876 | Meleagris_gallopavo |
| ENSG00000100201 | DDX17 | 98 | 67.273 | ENSMAUG00000020476 | Ddx17 | 86 | 88.827 | Mesocricetus_auratus |
| ENSG00000100201 | DDX17 | 100 | 99.042 | ENSMICG00000011601 | DDX17 | 100 | 99.042 | Microcebus_murinus |
| ENSG00000100201 | DDX17 | 98 | 95.119 | ENSMOCG00000010769 | Ddx17 | 100 | 95.119 | Microtus_ochrogaster |
| ENSG00000100201 | DDX17 | 73 | 95.122 | ENSMMOG00000007416 | si:dkey-156n14.5 | 89 | 87.664 | Mola_mola |
| ENSG00000100201 | DDX17 | 80 | 91.111 | ENSMMOG00000002806 | - | 91 | 88.594 | Mola_mola |
| ENSG00000100201 | DDX17 | 83 | 97.521 | ENSMODG00000009681 | DDX17 | 97 | 97.521 | Monodelphis_domestica |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSMALG00000002485 | si:dkey-156n14.5 | 95 | 72.981 | Monopterus_albus |
| ENSG00000100201 | DDX17 | 89 | 98.773 | MGP_CAROLIEiJ_G0020017 | Ddx17 | 100 | 98.773 | Mus_caroli |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSMUSG00000055065 | Ddx17 | 100 | 98.773 | Mus_musculus |
| ENSG00000100201 | DDX17 | 89 | 98.466 | MGP_PahariEiJ_G0020019 | Ddx17 | 100 | 98.466 | Mus_pahari |
| ENSG00000100201 | DDX17 | 89 | 98.773 | MGP_SPRETEiJ_G0020915 | Ddx17 | 100 | 98.773 | Mus_spretus |
| ENSG00000100201 | DDX17 | 89 | 99.846 | ENSMPUG00000013125 | DDX17 | 100 | 99.846 | Mustela_putorius_furo |
| ENSG00000100201 | DDX17 | 82 | 95.522 | ENSMLUG00000013151 | DDX17 | 91 | 95.462 | Myotis_lucifugus |
| ENSG00000100201 | DDX17 | 89 | 98.773 | ENSNGAG00000004001 | Ddx17 | 100 | 98.773 | Nannospalax_galili |
| ENSG00000100201 | DDX17 | 73 | 97.561 | ENSNBRG00000002592 | DDX17 | 87 | 89.005 | Neolamprologus_brichardi |
| ENSG00000100201 | DDX17 | 73 | 97.561 | ENSNBRG00000020059 | - | 93 | 86.329 | Neolamprologus_brichardi |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSNLEG00000015117 | DDX17 | 99 | 100.000 | Nomascus_leucogenys |
| ENSG00000100201 | DDX17 | 87 | 89.100 | ENSMEUG00000009627 | DDX17 | 100 | 89.100 | Notamacropus_eugenii |
| ENSG00000100201 | DDX17 | 100 | 98.765 | ENSOPRG00000014734 | DDX17 | 100 | 98.765 | Ochotona_princeps |
| ENSG00000100201 | DDX17 | 99 | 95.885 | ENSODEG00000013118 | DDX17 | 100 | 95.885 | Octodon_degus |
| ENSG00000100201 | DDX17 | 73 | 97.561 | ENSONIG00000018880 | si:dkey-156n14.5 | 91 | 89.390 | Oreochromis_niloticus |
| ENSG00000100201 | DDX17 | 89 | 99.385 | ENSOCUG00000004982 | DDX17 | 100 | 99.385 | Oryctolagus_cuniculus |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSORLG00000013685 | si:dkey-156n14.5 | 93 | 73.132 | Oryzias_latipes |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSORLG00020021743 | si:dkey-156n14.5 | 93 | 72.973 | Oryzias_latipes_hni |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSORLG00015022548 | si:dkey-156n14.5 | 93 | 73.291 | Oryzias_latipes_hsok |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSOMEG00000009515 | si:dkey-156n14.5 | 96 | 74.086 | Oryzias_melastigma |
| ENSG00000100201 | DDX17 | 89 | 99.387 | ENSOGAG00000001539 | DDX17 | 100 | 99.387 | Otolemur_garnettii |
| ENSG00000100201 | DDX17 | 87 | 99.684 | ENSOARG00000015489 | DDX17 | 100 | 99.684 | Ovis_aries |
| ENSG00000100201 | DDX17 | 100 | 100.000 | ENSPPAG00000042301 | DDX17 | 100 | 100.000 | Pan_paniscus |
| ENSG00000100201 | DDX17 | 100 | 99.316 | ENSPPRG00000008444 | DDX17 | 97 | 99.316 | Panthera_pardus |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSPTIG00000008733 | DDX17 | 99 | 93.568 | Panthera_tigris_altaica |
| ENSG00000100201 | DDX17 | 100 | 100.000 | ENSPTRG00000014369 | DDX17 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000100201 | DDX17 | 100 | 99.181 | ENSPANG00000015143 | DDX17 | 100 | 99.181 | Papio_anubis |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSPKIG00000002575 | si:dkey-156n14.5 | 94 | 75.583 | Paramormyrops_kingsleyae |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSPKIG00000007831 | DDX17 | 94 | 72.727 | Paramormyrops_kingsleyae |
| ENSG00000100201 | DDX17 | 89 | 98.160 | ENSPEMG00000021115 | Ddx17 | 100 | 98.160 | Peromyscus_maniculatus_bairdii |
| ENSG00000100201 | DDX17 | 89 | 98.000 | ENSPCIG00000001652 | DDX17 | 100 | 98.000 | Phascolarctos_cinereus |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSPREG00000000506 | si:dkey-156n14.5 | 92 | 74.522 | Poecilia_reticulata |
| ENSG00000100201 | DDX17 | 89 | 100.000 | ENSPPYG00000011821 | DDX17 | 100 | 100.000 | Pongo_abelii |
| ENSG00000100201 | DDX17 | 99 | 97.787 | ENSPCAG00000011672 | DDX17 | 100 | 97.787 | Procavia_capensis |
| ENSG00000100201 | DDX17 | 100 | 99.179 | ENSPCOG00000014360 | DDX17 | 100 | 99.179 | Propithecus_coquereli |
| ENSG00000100201 | DDX17 | 100 | 99.314 | ENSPVAG00000010133 | DDX17 | 100 | 99.314 | Pteropus_vampyrus |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSPNYG00000019669 | - | 94 | 74.878 | Pundamilia_nyererei |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSPNYG00000005694 | si:dkey-156n14.5 | 93 | 73.594 | Pundamilia_nyererei |
| ENSG00000100201 | DDX17 | 73 | 97.561 | ENSPNAG00000029948 | si:dkey-156n14.5 | 91 | 90.451 | Pygocentrus_nattereri |
| ENSG00000100201 | DDX17 | 89 | 99.080 | ENSRNOG00000051170 | Ddx17 | 100 | 99.080 | Rattus_norvegicus |
| ENSG00000100201 | DDX17 | 100 | 99.863 | ENSRBIG00000031437 | DDX17 | 100 | 99.863 | Rhinopithecus_bieti |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSRROG00000045378 | DDX17 | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000100201 | DDX17 | 71 | 70.000 | YNL112W | - | 81 | 61.451 | Saccharomyces_cerevisiae |
| ENSG00000100201 | DDX17 | 100 | 91.929 | ENSSBOG00000024547 | DDX17 | 100 | 91.929 | Saimiri_boliviensis_boliviensis |
| ENSG00000100201 | DDX17 | 87 | 97.950 | ENSSHAG00000004578 | DDX17 | 100 | 97.950 | Sarcophilus_harrisii |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSSFOG00015012308 | ddx17 | 95 | 74.807 | Scleropages_formosus |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSSMAG00000005509 | DDX17 | 94 | 75.845 | Scophthalmus_maximus |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSSMAG00000007794 | si:dkey-156n14.5 | 93 | 70.382 | Scophthalmus_maximus |
| ENSG00000100201 | DDX17 | 75 | 95.238 | ENSSPAG00000019808 | - | 93 | 89.390 | Stegastes_partitus |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSSPAG00000017130 | si:dkey-156n14.5 | 93 | 74.722 | Stegastes_partitus |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSSSCG00000000104 | DDX17 | 100 | 99.692 | Sus_scrofa |
| ENSG00000100201 | DDX17 | 87 | 89.515 | ENSTGUG00000010391 | DDX17 | 95 | 88.567 | Taeniopygia_guttata |
| ENSG00000100201 | DDX17 | 71 | 95.000 | ENSTRUG00000021931 | si:dkey-156n14.5 | 93 | 73.760 | Takifugu_rubripes |
| ENSG00000100201 | DDX17 | 87 | 98.420 | ENSTTRG00000005623 | DDX17 | 100 | 98.420 | Tursiops_truncatus |
| ENSG00000100201 | DDX17 | 100 | 96.990 | ENSUAMG00000007290 | DDX17 | 100 | 96.990 | Ursus_americanus |
| ENSG00000100201 | DDX17 | 100 | 92.593 | ENSVPAG00000001476 | DDX17 | 100 | 92.593 | Vicugna_pacos |
| ENSG00000100201 | DDX17 | 98 | 100.000 | ENSVVUG00000022762 | DDX17 | 96 | 100.000 | Vulpes_vulpes |
| ENSG00000100201 | DDX17 | 71 | 92.500 | ENSXETG00000006900 | ddx17 | 94 | 78.317 | Xenopus_tropicalis |
| ENSG00000100201 | DDX17 | 71 | 97.500 | ENSXMAG00000005078 | si:dkey-156n14.5 | 92 | 74.363 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000380 | alternative mRNA splicing, via spliceosome | 24275493.24910439. | IMP | Process |
| GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 23022728. | IMP | Process |
| GO:0001837 | epithelial to mesenchymal transition | 24910439. | IMP | Process |
| GO:0003713 | transcription coactivator activity | 17226766. | IDA | Function |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0003724 | RNA helicase activity | 8871553. | TAS | Function |
| GO:0005515 | protein binding | 14559993.15298701.15660129.16189514.18279852.18334637.21876179.22266867.22365833.22407013.24581491.24910439.24965446.25416956.26420826.26871637. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | 24910439. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005730 | nucleolus | - | IEA | Component |
| GO:0005829 | cytosol | - | IEA | Component |
| GO:0006357 | regulation of transcription by RNA polymerase II | 24275493. | IMP | Process |
| GO:0006364 | rRNA processing | - | IEA | Process |
| GO:0006396 | RNA processing | 8871553. | TAS | Process |
| GO:0008186 | RNA-dependent ATPase activity | 8871553. | TAS | Function |
| GO:0010586 | miRNA metabolic process | 27478153. | IMP | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016607 | nuclear speck | - | IDA | Component |
| GO:0030520 | intracellular estrogen receptor signaling pathway | 24275493. | IMP | Process |
| GO:0030521 | androgen receptor signaling pathway | 24275493. | IMP | Process |
| GO:0031047 | gene silencing by RNA | - | IEA | Process |
| GO:0045445 | myoblast differentiation | - | ISS | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 17226766. | IDA | Process |
| GO:0051607 | defense response to virus | - | IEA | Process |
| GO:2001014 | regulation of skeletal muscle cell differentiation | 17011493. | IMP | Process |