
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CHOL | |||
| ESCA | |||
| LUAD | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| SARC | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| SKCM | ||
| PRAD | ||
| KIRP | ||
| PAAD | ||
| PCPG | ||
| BLCA | ||
| CESC | ||
| LAML | ||
| DLBC | ||
| LGG | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000497339 | TELO2-202 | 1667 | - | ENSP00000456383 | 426 (aa) | - | H3BRS3 |
| ENST00000262319 | TELO2-201 | 3314 | XM_011522773 | ENSP00000262319 | 837 (aa) | XP_011521075 | Q9Y4R8 |
| ENST00000568240 | TELO2-207 | 1327 | - | - | - (aa) | - | - |
| ENST00000569744 | TELO2-208 | 575 | - | - | - (aa) | - | - |
| ENST00000567427 | TELO2-206 | 634 | - | - | - (aa) | - | - |
| ENST00000564507 | TELO2-204 | 903 | - | - | - (aa) | - | - |
| ENST00000563676 | TELO2-203 | 555 | - | - | - (aa) | - | - |
| ENST00000567423 | TELO2-205 | 479 | - | ENSP00000457461 | 160 (aa) | - | H3BU45 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000100726 | Platelet Function Tests | 2.7134300E-005 | - |
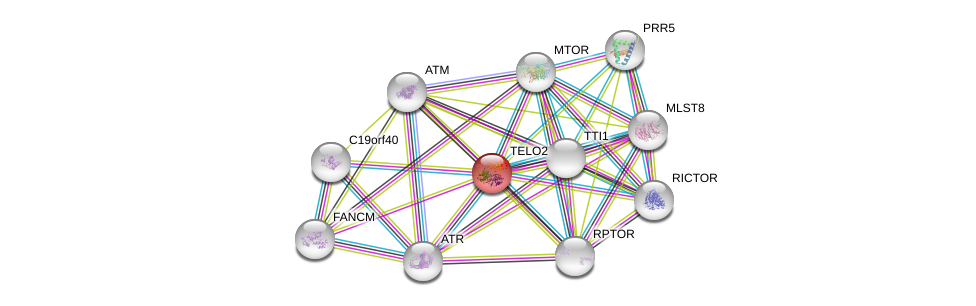
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000100726 | TELO2 | 100 | 75.060 | ENSMUSG00000024170 | Telo2 | 99 | 75.060 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000723 | telomere maintenance | 21873635. | IBA | Process |
| GO:0000784 | nuclear chromosome, telomeric region | 21873635. | IBA | Component |
| GO:0005515 | protein binding | 18160036.20371770.20801936.20810650.20864032.23263282.24036451.24656813. | IPI | Function |
| GO:0005634 | nucleus | 20864032. | IDA | Component |
| GO:0005737 | cytoplasm | 20864032.23263282. | IDA | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0016020 | membrane | - | IEA | Component |
| GO:0016604 | nuclear body | - | IDA | Component |
| GO:0019901 | protein kinase binding | 20864032. | IPI | Function |
| GO:0031931 | TORC1 complex | 23263282. | IDA | Component |
| GO:0031932 | TORC2 complex | 23263282. | IDA | Component |
| GO:0032006 | regulation of TOR signaling | 23263282. | IMP | Process |
| GO:0034399 | nuclear periphery | 20864032. | IDA | Component |
| GO:0042162 | telomeric DNA binding | 21873635. | IBA | Function |
| GO:0044877 | protein-containing complex binding | 20864032. | IDA | Function |
| GO:0050821 | protein stabilization | 24036451. | IMP | Process |
| GO:0051879 | Hsp90 protein binding | 20864032. | IPI | Function |
| GO:0060090 | molecular adaptor activity | - | IEA | Function |
| GO:0070209 | ASTRA complex | 21873635. | IBA | Component |
| GO:0071902 | positive regulation of protein serine/threonine kinase activity | 24036451. | IMP | Process |
| GO:1904263 | positive regulation of TORC1 signaling | 24036451. | IMP | Process |
| GO:1904515 | positive regulation of TORC2 signaling | 24036451. | IMP | Process |