
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 27495250 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THCA | |||||||
| THCA | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CHOL | |||
| KIRP | |||
| PAAD |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| SARC | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| BLCA | ||
| CESC | ||
| LUAD | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000555363 | SEC23A-209 | 704 | - | - | - (aa) | - | - |
| ENST00000557280 | SEC23A-213 | 561 | - | ENSP00000452575 | 85 (aa) | - | G3V5X8 |
| ENST00000553970 | SEC23A-206 | 574 | - | ENSP00000451924 | 86 (aa) | - | G3V4Q2 |
| ENST00000537403 | SEC23A-202 | 4215 | - | ENSP00000444193 | 563 (aa) | - | Q15436 |
| ENST00000545328 | SEC23A-203 | 2734 | - | ENSP00000445393 | 736 (aa) | - | F5H365 |
| ENST00000625395 | SEC23A-215 | 261 | - | ENSP00000486142 | 86 (aa) | - | G3V4Q2 |
| ENST00000556092 | SEC23A-212 | 571 | - | ENSP00000451230 | 140 (aa) | - | G3V3G5 |
| ENST00000557437 | SEC23A-214 | 575 | - | ENSP00000452390 | 40 (aa) | - | G3V5K1 |
| ENST00000548032 | SEC23A-204 | 907 | - | ENSP00000447489 | 108 (aa) | - | G3V1W4 |
| ENST00000555017 | SEC23A-208 | 588 | - | ENSP00000450819 | 169 (aa) | - | G3V2R6 |
| ENST00000555682 | SEC23A-211 | 398 | - | - | - (aa) | - | - |
| ENST00000555425 | SEC23A-210 | 547 | - | ENSP00000451999 | 93 (aa) | - | G3V4V1 |
| ENST00000307712 | SEC23A-201 | 4135 | XM_005267262 | ENSP00000306881 | 765 (aa) | XP_005267319 | Q15436 |
| ENST00000554615 | SEC23A-207 | 3813 | - | - | - (aa) | - | - |
| ENST00000553925 | SEC23A-205 | 484 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04141 | Protein processing in endoplasmic reticulum | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000100934 | Taste Perception | 1.8140000E-006 | - |
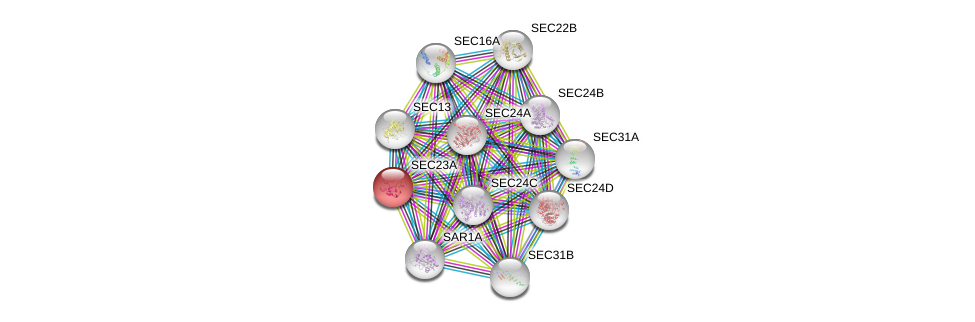
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000100934 | SEC23A | 99 | 81.522 | FBgn0262125 | Sec23 | 99 | 74.065 | Drosophila_melanogaster |
| ENSG00000100934 | SEC23A | 100 | 92.500 | ENSG00000101310 | SEC23B | 100 | 96.875 | Homo_sapiens |
| ENSG00000100934 | SEC23A | 99 | 49.677 | YPR181C | 99 | 49.677 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000139 | Golgi membrane | - | IEA | Component |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | - | TAS | Process |
| GO:0005096 | GTPase activator activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 10075675.16091426.17499046.18692470.20946353.27018634.27551091.28442536. | IPI | Function |
| GO:0005789 | endoplasmic reticulum membrane | - | TAS | Component |
| GO:0005829 | cytosol | 8898360. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006886 | intracellular protein transport | - | IEA | Process |
| GO:0006888 | ER to Golgi vesicle-mediated transport | - | TAS | Process |
| GO:0008270 | zinc ion binding | 18843296. | IDA | Function |
| GO:0012507 | ER to Golgi transport vesicle membrane | 8898360. | IGI | Component |
| GO:0012507 | ER to Golgi transport vesicle membrane | - | TAS | Component |
| GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II | - | TAS | Process |
| GO:0019898 | extrinsic component of membrane | 8898360. | IDA | Component |
| GO:0030127 | COPII vesicle coat | 21873635. | IBA | Component |
| GO:0030127 | COPII vesicle coat | 17499046. | IDA | Component |
| GO:0043547 | positive regulation of GTPase activity | - | IEA | Process |
| GO:0048208 | COPII vesicle coating | - | TAS | Process |
| GO:0048471 | perinuclear region of cytoplasm | - | IEA | Component |
| GO:0070971 | endoplasmic reticulum exit site | 21873635. | IBA | Component |
| GO:0070971 | endoplasmic reticulum exit site | 8898360.28442536. | IDA | Component |
| GO:0072659 | protein localization to plasma membrane | - | IEA | Process |
| GO:0090110 | cargo loading into COPII-coated vesicle | 21873635. | IBA | Process |
| GO:0090110 | cargo loading into COPII-coated vesicle | 17499046.18843296. | IDA | Process |
| GO:0090110 | cargo loading into COPII-coated vesicle | 8898360. | IGI | Process |