
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| LAML |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| SARC | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| SKCM | ||
| ESCA | ||
| CESC | ||
| LAML | ||
| UCEC | ||
| LGG | ||
| LUAD | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000646673 | SAMHD1-221 | 5298 | - | ENSP00000493536 | 626 (aa) | - | Q9Y3Z3 |
| ENST00000644114 | SAMHD1-213 | 1735 | - | ENSP00000494354 | 413 (aa) | - | A0A2R8YD90 |
| ENST00000646066 | SAMHD1-219 | 1671 | - | ENSP00000495432 | 556 (aa) | - | Q9Y3Z3 |
| ENST00000643907 | SAMHD1-211 | 540 | - | - | - (aa) | - | - |
| ENST00000644688 | SAMHD1-216 | 951 | - | - | - (aa) | - | - |
| ENST00000647095 | SAMHD1-225 | 5492 | - | - | - (aa) | - | - |
| ENST00000643003 | SAMHD1-206 | 546 | - | - | - (aa) | - | - |
| ENST00000643161 | SAMHD1-208 | 816 | - | - | - (aa) | - | - |
| ENST00000646866 | SAMHD1-222 | 975 | - | ENSP00000495737 | 129 (aa) | - | A0A2R8Y755 |
| ENST00000262878 | SAMHD1-201 | 4334 | XM_005260384 | ENSP00000262878 | 591 (aa) | XP_005260441 | Q9Y3Z3 |
| ENST00000646904 | SAMHD1-224 | 4672 | - | ENSP00000494823 | 285 (aa) | - | A0A2R8YF51 |
| ENST00000646869 | SAMHD1-223 | 3209 | - | ENSP00000495667 | 626 (aa) | - | Q9Y3Z3 |
| ENST00000646121 | SAMHD1-220 | 643 | - | ENSP00000495645 | 89 (aa) | - | A0A2R8YG48 |
| ENST00000645444 | SAMHD1-218 | 3763 | - | ENSP00000495381 | 212 (aa) | - | A0A2R8Y6J9 |
| ENST00000645033 | SAMHD1-217 | 4680 | - | ENSP00000494520 | 289 (aa) | - | A0A2R8Y5D2 |
| ENST00000643243 | SAMHD1-209 | 1260 | - | - | - (aa) | - | - |
| ENST00000465985 | SAMHD1-202 | 513 | - | ENSP00000496075 | 104 (aa) | - | A0A2R8YF18 |
| ENST00000644250 | SAMHD1-214 | 1824 | - | ENSP00000493810 | 44 (aa) | - | A0A2R8YDI8 |
| ENST00000647459 | SAMHD1-227 | 3146 | - | - | - (aa) | - | - |
| ENST00000644370 | SAMHD1-215 | 527 | - | - | - (aa) | - | - |
| ENST00000643918 | SAMHD1-212 | 2967 | XM_011528761 | ENSP00000493928 | 589 (aa) | XP_011527063 | A0A2R8YCS7 |
| ENST00000642246 | SAMHD1-204 | 4508 | - | ENSP00000494979 | 127 (aa) | - | A0A2R8Y7R5 |
| ENST00000642186 | SAMHD1-203 | 3312 | - | ENSP00000494436 | 289 (aa) | - | A0A2R8Y5D2 |
| ENST00000647163 | SAMHD1-226 | 3440 | - | ENSP00000494313 | 289 (aa) | - | A0A2R8Y5D2 |
| ENST00000642616 | SAMHD1-205 | 1424 | - | ENSP00000494271 | 156 (aa) | - | A0A2R8Y586 |
| ENST00000643078 | SAMHD1-207 | 1395 | - | ENSP00000496474 | 127 (aa) | - | A0A2R8Y7R5 |
| ENST00000643825 | SAMHD1-210 | 522 | - | ENSP00000495448 | 139 (aa) | - | A0A2R8YFV6 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa05170 | Human immunodeficiency virus 1 infection | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000101347 | Platelet Function Tests | 7.5755000E-005 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000101347 | rs11696169 | 20 | 36923214 | C | Low tan response | 29739929 | [0.088-0.284] unit increase | 0.186 | EFO_0004279 |
| ENSG00000101347 | rs6030369 | 20 | 36949294 | ? | Hair color | 30595370 | EFO_0007822 |
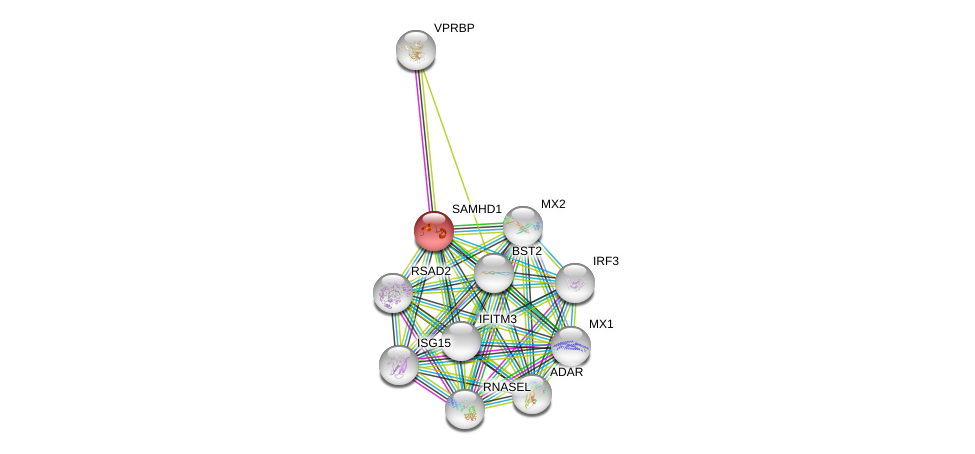
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000724 | double-strand break repair via homologous recombination | 28834754.29670289. | IDA | Process |
| GO:0003676 | nucleic acid binding | 22461318. | IDA | Function |
| GO:0003697 | single-stranded DNA binding | 29670289. | IDA | Function |
| GO:0003723 | RNA binding | 22461318. | IDA | Function |
| GO:0004540 | ribonuclease activity | 25038827. | IDA | Function |
| GO:0004540 | ribonuclease activity | 29670289. | IDA | Function |
| GO:0005515 | protein binding | 21903422.23414517.23601106.28834754.28871089.29670289. | IPI | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 19525956. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005886 | plasma membrane | - | IDA | Component |
| GO:0006203 | dGTP catabolic process | 21873635. | IBA | Process |
| GO:0006203 | dGTP catabolic process | 24217394. | IDA | Process |
| GO:0006955 | immune response | 11064105. | NAS | Process |
| GO:0006974 | cellular response to DNA damage stimulus | 28834754.29670289. | IDA | Process |
| GO:0008270 | zinc ion binding | 24217394. | IDA | Function |
| GO:0008832 | dGTPase activity | 21873635. | IBA | Function |
| GO:0008832 | dGTPase activity | 23601106.24217394.26294762. | IDA | Function |
| GO:0009264 | deoxyribonucleotide catabolic process | 23601106.26294762. | IDA | Process |
| GO:0009264 | deoxyribonucleotide catabolic process | - | TAS | Process |
| GO:0016446 | somatic hypermutation of immunoglobulin genes | - | ISS | Process |
| GO:0016793 | triphosphoric monoester hydrolase activity | 23601106.26294762. | IDA | Function |
| GO:0032567 | dGTP binding | 24141705. | IDA | Function |
| GO:0035861 | site of double-strand break | 28834754.29670289. | IDA | Component |
| GO:0042802 | identical protein binding | 22056990.24141705. | IPI | Function |
| GO:0045088 | regulation of innate immune response | 21873635. | IBA | Process |
| GO:0045088 | regulation of innate immune response | 29670289. | IDA | Process |
| GO:0045088 | regulation of innate immune response | 19525956. | IMP | Process |
| GO:0046061 | dATP catabolic process | 24141705. | IDA | Process |
| GO:0051289 | protein homotetramerization | 23601106.24217394.26294762. | IDA | Process |
| GO:0051607 | defense response to virus | 21873635. | IBA | Process |
| GO:0051607 | defense response to virus | 23601106.26294762. | IDA | Process |
| GO:0051607 | defense response to virus | 19525956. | IMP | Process |
| GO:0060337 | type I interferon signaling pathway | - | TAS | Process |
| GO:0060339 | negative regulation of type I interferon-mediated signaling pathway | 29670289. | IDA | Process |
| GO:0090501 | RNA phosphodiester bond hydrolysis | - | IEA | Process |
| GO:0097197 | tetraspanin-enriched microdomain | 28871089. | IDA | Component |
| GO:0110025 | DNA strand resection involved in replication fork processing | 28834754.29670289. | IDA | Process |