
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRC | |||
| PRAD | |||
| SARC | |||
| TGCT | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| ACC | ||
| SKCM | ||
| BRCA | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| LAML | ||
| UCEC | ||
| LUAD |
| PID | Title | Method | Time | Author | Doi |
|---|---|---|---|---|---|
| 30352994 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000358397 | ESRP1-201 | 3756 | - | ENSP00000351168 | 677 (aa) | - | Q6NXG1 |
| ENST00000522756 | ESRP1-209 | 609 | - | ENSP00000428490 | 40 (aa) | - | E5RI26 |
| ENST00000522920 | ESRP1-210 | 402 | - | - | - (aa) | - | - |
| ENST00000642581 | ESRP1-212 | 3782 | - | ENSP00000495733 | 681 (aa) | - | Q6NXG1 |
| ENST00000646773 | ESRP1-213 | 3274 | - | ENSP00000494213 | 608 (aa) | - | Q6NXG1 |
| ENST00000433389 | ESRP1-203 | 3806 | XM_005250992 | ENSP00000405738 | 681 (aa) | XP_005251049 | Q6NXG1 |
| ENST00000519505 | ESRP1-207 | 3068 | - | ENSP00000430118 | 502 (aa) | - | H0YBR2 |
| ENST00000423620 | ESRP1-202 | 3746 | XM_005250991 | ENSP00000407349 | 659 (aa) | XP_005251048 | Q6NXG1 |
| ENST00000517610 | ESRP1-206 | 2139 | - | ENSP00000429125 | 522 (aa) | - | H0YBB3 |
| ENST00000520385 | ESRP1-208 | 764 | - | - | - (aa) | - | - |
| ENST00000517556 | ESRP1-205 | 457 | - | - | - (aa) | - | - |
| ENST00000454170 | ESRP1-204 | 3465 | - | ENSP00000402766 | 604 (aa) | - | A0A2U3TZN9 |
| ENST00000523347 | ESRP1-211 | 1813 | - | - | - (aa) | - | - |
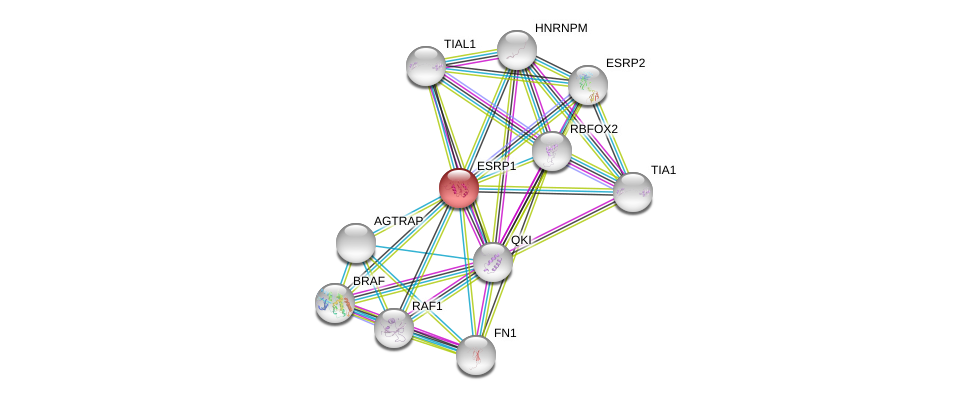
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000104413 | ESRP1 | 100 | 97.269 | ENSMUSG00000040728 | Esrp1 | 100 | 97.269 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003729 | mRNA binding | 19285943. | IDA | Function |
| GO:0005515 | protein binding | 25416956. | IPI | Function |
| GO:0005634 | nucleus | 19285943. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0006397 | mRNA processing | - | IEA | Process |
| GO:0008380 | RNA splicing | - | IEA | Process |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | - | TAS | Process |
| GO:0016604 | nuclear body | - | IDA | Component |
| GO:0042669 | regulation of inner ear auditory receptor cell fate specification | - | ISS | Process |
| GO:0043484 | regulation of RNA splicing | 19285943. | IDA | Process |