
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS | |||||||
| UCS | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| DLBC | |||
| ESCA | |||
| PAAD | |||
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| ACC | ||
| SKCM | ||
| PRAD | ||
| KIRP | ||
| COAD | ||
| CESC | ||
| KICH | ||
| LIHC | ||
| LGG |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000221130 | GSR-201 | 3192 | - | ENSP00000221130 | 522 (aa) | - | P00390 |
| ENST00000537535 | GSR-204 | 1323 | - | ENSP00000438845 | 440 (aa) | - | P00390 |
| ENST00000643653 | GSR-208 | 2804 | - | ENSP00000494854 | 123 (aa) | - | A0A2R8YF59 |
| ENST00000541648 | GSR-205 | 1410 | - | ENSP00000444559 | 469 (aa) | - | P00390 |
| ENST00000643525 | GSR-207 | 1802 | - | - | - (aa) | - | - |
| ENST00000546342 | GSR-206 | 1482 | - | ENSP00000445516 | 493 (aa) | - | P00390 |
| ENST00000521479 | GSR-202 | 582 | - | - | - (aa) | - | - |
| ENST00000523295 | GSR-203 | 913 | - | ENSP00000431044 | 131 (aa) | - | H0YC68 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000104687 | Arteries | 1.5140000E-006 | - |
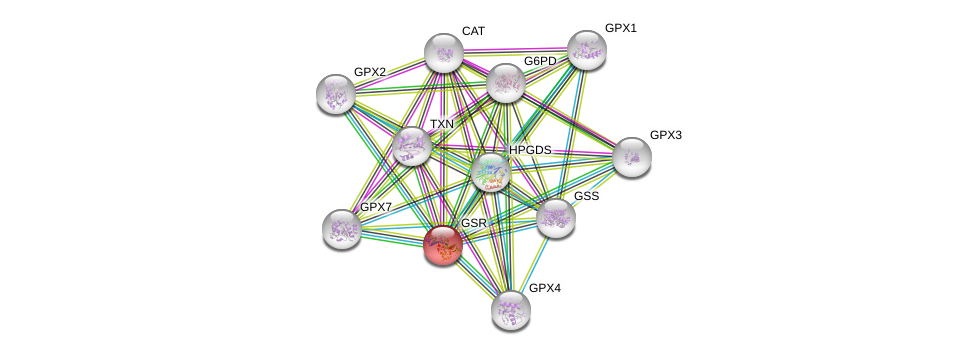
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000104687 | GSR | 85 | 56.410 | YPL091W | GLR1 | 95 | 50.740 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004362 | glutathione-disulfide reductase activity | 21873635. | IBA | Function |
| GO:0005739 | mitochondrion | 21873635. | IBA | Component |
| GO:0005759 | mitochondrial matrix | - | TAS | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006749 | glutathione metabolic process | 21873635. | IBA | Process |
| GO:0009055 | electron transfer activity | 947404. | TAS | Function |
| GO:0009897 | external side of plasma membrane | - | IEA | Component |
| GO:0015949 | nucleobase-containing small molecule interconversion | - | TAS | Process |
| GO:0022900 | electron transport chain | - | IEA | Process |
| GO:0034599 | cellular response to oxidative stress | 21873635. | IBA | Process |
| GO:0034599 | cellular response to oxidative stress | - | TAS | Process |
| GO:0045454 | cell redox homeostasis | 21873635. | IBA | Process |
| GO:0050660 | flavin adenine dinucleotide binding | 21873635. | IBA | Function |
| GO:0050661 | NADP binding | - | IEA | Function |
| GO:0070062 | extracellular exosome | 19056867.23533145. | HDA | Component |
| GO:0098869 | cellular oxidant detoxification | - | IEA | Process |