
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CHOL | |||
| COAD | |||
| LGG | |||
| LIHC | |||
| MESO | |||
| PAAD | |||
| READ | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| SARC | ||
| ACC | ||
| UCS | ||
| HNSC | ||
| SKCM | ||
| PRAD | ||
| ESCA | ||
| KIRP | ||
| CESC | ||
| UCEC | ||
| LIHC | ||
| CHOL | ||
| UVM | ||
| OV |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 17135243 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000278100 | BCCIP-201 | 1236 | - | ENSP00000278100 | 314 (aa) | - | Q9P287 |
| ENST00000478798 | BCCIP-205 | 1461 | - | - | - (aa) | - | - |
| ENST00000368759 | BCCIP-203 | 1433 | - | ENSP00000357748 | 322 (aa) | - | Q9P287 |
| ENST00000463330 | BCCIP-204 | 500 | - | - | - (aa) | - | - |
| ENST00000299130 | BCCIP-202 | 2337 | - | ENSP00000299130 | 292 (aa) | - | Q9P287 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000107949 | rs11244669 | 10 | 125844213 | ? | Red cell distribution width | 30595370 | EFO_0005192 | ||
| ENSG00000107949 | rs11244669 | 10 | 125844213 | ? | Mean corpuscular hemoglobin | 30595370 | EFO_0004527 | ||
| ENSG00000107949 | rs11244669 | 10 | 125844213 | ? | Red blood cell count | 30595370 | EFO_0004305 |
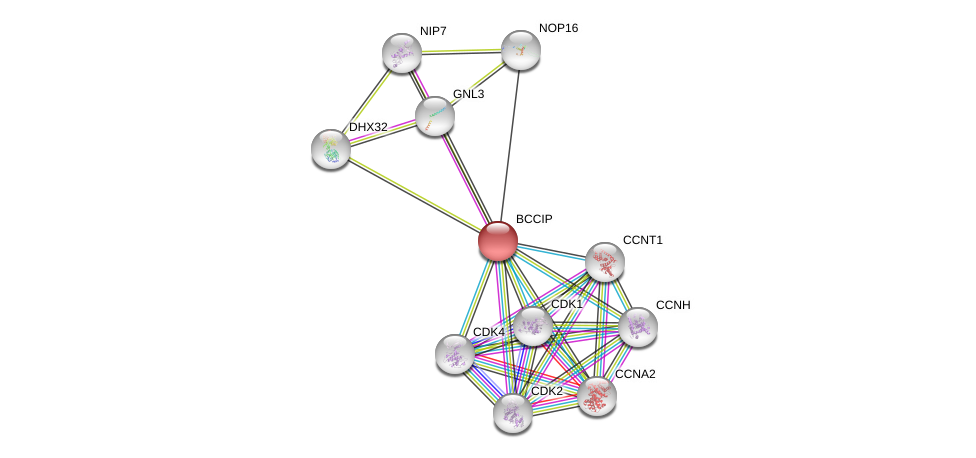
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000055 | ribosomal large subunit export from nucleus | 21873635. | IBA | Process |
| GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 21873635. | IBA | Process |
| GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 10878006. | IDA | Process |
| GO:0000132 | establishment of mitotic spindle orientation | 21873635. | IBA | Process |
| GO:0000226 | microtubule cytoskeleton organization | 28394342. | IMP | Process |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0005515 | protein binding | 16189514.18440304.25416956.28931820. | IPI | Function |
| GO:0005634 | nucleus | 10878006. | TAS | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005813 | centrosome | 21873635. | IBA | Component |
| GO:0005813 | centrosome | 28394342. | IDA | Component |
| GO:0005814 | centriole | 21873635. | IBA | Component |
| GO:0005814 | centriole | 28394342. | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0006281 | DNA repair | - | IEA | Process |
| GO:0006974 | cellular response to DNA damage stimulus | 21873635. | IBA | Process |
| GO:0007052 | mitotic spindle organization | 21873635. | IBA | Process |
| GO:0007052 | mitotic spindle organization | 28394342. | IMP | Process |
| GO:0015631 | tubulin binding | 21873635. | IBA | Function |
| GO:0019207 | kinase regulator activity | 21873635. | IBA | Function |
| GO:0019207 | kinase regulator activity | 10878006. | IDA | Function |
| GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex | 21873635. | IBA | Component |
| GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex | 10878006. | IDA | Component |
| GO:0034453 | microtubule anchoring | 21873635. | IBA | Process |
| GO:0034453 | microtubule anchoring | 28394342. | IMP | Process |
| GO:0061101 | neuroendocrine cell differentiation | 18440304. | IDA | Process |
| GO:0090307 | mitotic spindle assembly | 21873635. | IBA | Process |
| GO:0090307 | mitotic spindle assembly | 28394342. | IMP | Process |
| GO:0097431 | mitotic spindle pole | 21873635. | IBA | Component |
| GO:0097431 | mitotic spindle pole | 28394342. | IDA | Component |