
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| LAML | |||
| LIHC | |||
| LUAD | |||
| SKCM | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| SARC | ||
| ACC | ||
| HNSC | ||
| LAML | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| LUAD | ||
| UVM | ||
| OV |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 30718516 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000498643 | YWHAE-207 | 312 | - | - | - (aa) | - | - |
| ENST00000496706 | YWHAE-206 | 732 | - | ENSP00000466324 | 115 (aa) | - | K7EM20 |
| ENST00000469398 | YWHAE-203 | 676 | - | - | - (aa) | - | - |
| ENST00000575977 | YWHAE-212 | 540 | - | ENSP00000460712 | 107 (aa) | - | I3L3T1 |
| ENST00000576854 | YWHAE-213 | 1374 | - | - | - (aa) | - | - |
| ENST00000466227 | YWHAE-202 | 1377 | - | ENSP00000464883 | 109 (aa) | - | K7EIT4 |
| ENST00000571732 | YWHAE-208 | 1818 | XM_017025005 | ENSP00000461762 | 233 (aa) | XP_016880494 | P62258 |
| ENST00000573026 | YWHAE-210 | 780 | - | ENSP00000458386 | 38 (aa) | - | I3L0W5 |
| ENST00000489287 | YWHAE-205 | 465 | - | - | - (aa) | - | - |
| ENST00000572108 | YWHAE-209 | 258 | - | - | - (aa) | - | - |
| ENST00000486241 | YWHAE-204 | 691 | - | - | - (aa) | - | - |
| ENST00000264335 | YWHAE-201 | 2211 | XM_005256784 | ENSP00000264335 | 255 (aa) | XP_005256841 | P62258 |
| ENST00000573196 | YWHAE-211 | 739 | - | ENSP00000461766 | 94 (aa) | - | B4DJF2 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000108953 | rs9908552 | 17 | 1394697 | T | Schizophrenia | 26198764 | [NR] | 1.05 | EFO_0000692 |
| ENSG00000108953 | rs56007784 | 17 | 1387656 | C | Schizophrenia | 28991256 | [1.04-1.08] | 1.059 | EFO_0000692 |
| ENSG00000108953 | rs75543711 | 17 | 1352780 | ? | Reaction time | 29844566 | [0.0061-0.0128] unit decrease | 0.009456 | EFO_0008393 |
| ENSG00000108953 | rs56007784 | 17 | 1387656 | C | Schizophrenia | 30285260 | [1.03-1.07] | 1.052 | EFO_0000692 |
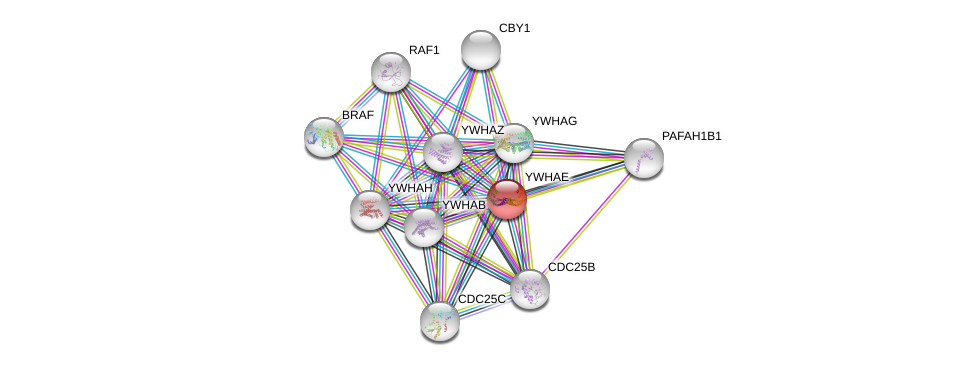
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000108953 | YWHAE | 99 | 71.930 | ENSG00000170027 | YWHAG | 100 | 63.968 | Homo_sapiens |
| ENSG00000108953 | YWHAE | 100 | 69.565 | ENSG00000128245 | YWHAH | 99 | 63.265 | Homo_sapiens |
| ENSG00000108953 | YWHAE | 100 | 100.000 | ENSMUSG00000020849 | Ywhae | 100 | 100.000 | Mus_musculus |
| ENSG00000108953 | YWHAE | 99 | 71.930 | ENSMUSG00000051391 | Ywhag | 100 | 63.968 | Mus_musculus |
| ENSG00000108953 | YWHAE | 100 | 69.565 | ENSMUSG00000018965 | Ywhah | 99 | 63.265 | Mus_musculus |
| ENSG00000108953 | YWHAE | 96 | 83.636 | YDR099W | BMH2 | 93 | 73.913 | Saccharomyces_cerevisiae |
| ENSG00000108953 | YWHAE | 96 | 84.545 | YER177W | BMH1 | 92 | 75.203 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000086 | G2/M transition of mitotic cell cycle | - | TAS | Process |
| GO:0000165 | MAPK cascade | 12917326. | IDA | Process |
| GO:0001764 | neuron migration | - | IEA | Process |
| GO:0003064 | regulation of heart rate by hormone | 11953308. | NAS | Process |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0005246 | calcium channel regulator activity | 18029012. | IDA | Function |
| GO:0005515 | protein binding | 1266503.7644510.10644344.11504882.11697890.12392720.12917326.14743216.15722337.16099986.16227609.16260042.16376338.16407301.16511560.16775625.17085597.18045992.18227151.18356162.18458160.18573912.19172738.19640509.20618440.20642453.20936779.21044950.21242966.21282530.21346153.21988832.22151054.22246185.23075850.23572552.23622247.23752268.24255178.24366813.24725412.24947832.25588844.25751139.25852190.26496610.27173435.29128334.29769719.30611118. | IPI | Function |
| GO:0005634 | nucleus | 12917326. | IDA | Component |
| GO:0005737 | cytoplasm | 12917326. | IDA | Component |
| GO:0005739 | mitochondrion | - | IEA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005871 | kinesin complex | - | IEA | Component |
| GO:0005886 | plasma membrane | 18029012. | IDA | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0006605 | protein targeting | - | IEA | Process |
| GO:0010389 | regulation of G2/M transition of mitotic cell cycle | - | TAS | Process |
| GO:0015459 | potassium channel regulator activity | 11953308. | IDA | Function |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0019899 | enzyme binding | 10788521. | IPI | Function |
| GO:0019904 | protein domain specific binding | - | IEA | Function |
| GO:0021762 | substantia nigra development | 22926577. | HEP | Process |
| GO:0021766 | hippocampus development | - | IEA | Process |
| GO:0021987 | cerebral cortex development | - | IEA | Process |
| GO:0023026 | MHC class II protein complex binding | 20458337. | HDA | Function |
| GO:0031625 | ubiquitin protein ligase binding | 19725078. | IPI | Function |
| GO:0034504 | protein localization to nucleus | 29769719. | IMP | Process |
| GO:0034605 | cellular response to heat | 12917326. | IDA | Process |
| GO:0035329 | hippo signaling | - | TAS | Process |
| GO:0035556 | intracellular signal transduction | 7644510. | TAS | Process |
| GO:0042470 | melanosome | - | IEA | Component |
| GO:0042802 | identical protein binding | 17085597.20936779. | IPI | Function |
| GO:0042826 | histone deacetylase binding | 10869435. | IPI | Function |
| GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | - | TAS | Process |
| GO:0044325 | ion channel binding | 11953308.18029012. | IPI | Function |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0046827 | positive regulation of protein export from nucleus | 12917326. | IDA | Process |
| GO:0046982 | protein heterodimerization activity | 11953308. | IPI | Function |
| GO:0050815 | phosphoserine residue binding | 10869435. | IPI | Function |
| GO:0051219 | phosphoprotein binding | 10869435. | IPI | Function |
| GO:0051480 | regulation of cytosolic calcium ion concentration | 18029012. | IDA | Process |
| GO:0060306 | regulation of membrane repolarization | 11953308. | IDA | Process |
| GO:0061024 | membrane organization | - | TAS | Process |
| GO:0070062 | extracellular exosome | 19056867.19199708.20458337.23533145. | HDA | Component |
| GO:0086013 | membrane repolarization during cardiac muscle cell action potential | 11953308. | IC | Process |
| GO:0086091 | regulation of heart rate by cardiac conduction | 11953308. | IC | Process |
| GO:0090724 | central region of growth cone | - | IEA | Component |
| GO:0097110 | scaffold protein binding | 10409742. | IPI | Function |
| GO:0097711 | ciliary basal body-plasma membrane docking | - | TAS | Process |
| GO:0098978 | glutamatergic synapse | - | IEA | Component |
| GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | - | IEA | Process |
| GO:1900034 | regulation of cellular response to heat | - | TAS | Process |
| GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | - | TAS | Process |
| GO:1901016 | regulation of potassium ion transmembrane transporter activity | 11953308. | IDA | Process |
| GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 18029012. | IDA | Process |
| GO:1902309 | negative regulation of peptidyl-serine dephosphorylation | 11953308. | IDA | Process |
| GO:1905913 | negative regulation of calcium ion export across plasma membrane | 18029012. | IDA | Process |