
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| THCA | |||
| THYM | |||
| UCEC | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| HNSC | ||
| LUSC | ||
| KIRP | ||
| COAD | ||
| CESC | ||
| LAML | ||
| UVM | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000395319 | ALDOC-202 | 1519 | - | ENSP00000378729 | 336 (aa) | - | A8MVZ9 |
| ENST00000395321 | ALDOC-203 | 1722 | XM_005257949 | ENSP00000378731 | 364 (aa) | XP_005258006 | P09972 |
| ENST00000578590 | ALDOC-206 | 839 | - | ENSP00000463118 | 122 (aa) | - | J3QKK1 |
| ENST00000460201 | ALDOC-205 | 922 | - | ENSP00000463174 | 180 (aa) | - | J3QKP5 |
| ENST00000581807 | ALDOC-207 | 580 | - | ENSP00000465623 | 135 (aa) | - | K7EKH5 |
| ENST00000226253 | ALDOC-201 | 1982 | - | ENSP00000226253 | 364 (aa) | - | P09972 |
| ENST00000584086 | ALDOC-209 | 686 | - | ENSP00000462674 | 175 (aa) | - | J3KSV6 |
| ENST00000435638 | ALDOC-204 | 579 | - | ENSP00000398976 | 149 (aa) | - | C9J8F3 |
| ENST00000582381 | ALDOC-208 | 571 | - | - | - (aa) | - | - |
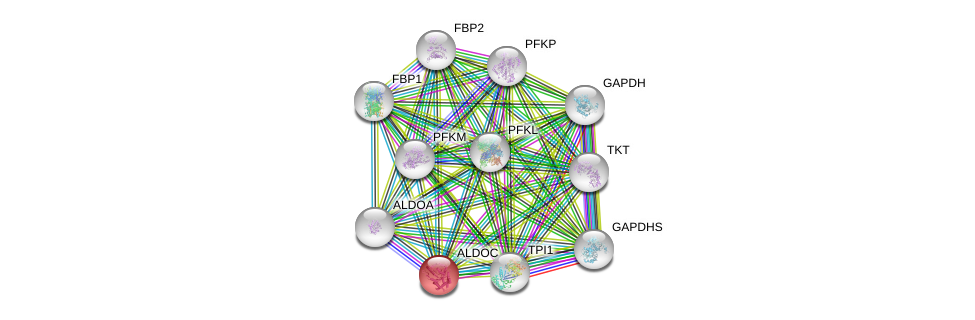
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000109107 | ALDOC | 100 | 67.407 | WBGene00011474 | aldo-1 | 98 | 65.084 | Caenorhabditis_elegans |
| ENSG00000109107 | ALDOC | 98 | 61.732 | WBGene00017166 | aldo-2 | 97 | 62.396 | Caenorhabditis_elegans |
| ENSG00000109107 | ALDOC | 100 | 83.333 | ENSG00000149925 | ALDOA | 100 | 85.784 | Homo_sapiens |
| ENSG00000109107 | ALDOC | 100 | 83.889 | ENSMUSG00000030695 | Aldoa | 100 | 87.712 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004332 | fructose-bisphosphate aldolase activity | 9244396.15537755. | IDA | Function |
| GO:0005515 | protein binding | 11399750.25416956.25910212. | IPI | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005856 | cytoskeleton | 9244396. | IC | Component |
| GO:0006000 | fructose metabolic process | 3105602. | TAS | Process |
| GO:0006094 | gluconeogenesis | - | TAS | Process |
| GO:0008092 | cytoskeletal protein binding | 9244396. | IDA | Function |
| GO:0030388 | fructose 1,6-bisphosphate metabolic process | 9244396.15537755. | IDA | Process |
| GO:0030855 | epithelial cell differentiation | 21492153. | IEP | Process |
| GO:0034774 | secretory granule lumen | - | TAS | Component |
| GO:0043312 | neutrophil degranulation | - | TAS | Process |
| GO:0061621 | canonical glycolysis | - | TAS | Process |
| GO:0070062 | extracellular exosome | 19056867.20458337.23533145. | HDA | Component |
| GO:1904724 | tertiary granule lumen | - | TAS | Component |
| GO:1904813 | ficolin-1-rich granule lumen | - | TAS | Component |