
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 27042257 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| BLCA | |||
| CHOL | |||
| DLBC | |||
| ESCA | |||
| KIRC | |||
| LUAD | |||
| MESO | |||
| PAAD | |||
| PCPG | |||
| READ | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| STAD | ||
| MESO | ||
| ACC | ||
| KIRP | ||
| PAAD | ||
| BLCA | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| CHOL | ||
| LUAD | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000264596 | zf-RanBP | PF00641.18 | 5.3e-06 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000264596 | NEIL3-201 | 2408 | XM_017008360 | ENSP00000264596 | 605 (aa) | XP_016863849 | Q8TAT5 |
| ENST00000513321 | NEIL3-202 | 1528 | - | ENSP00000424735 | 63 (aa) | - | D6RAV1 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
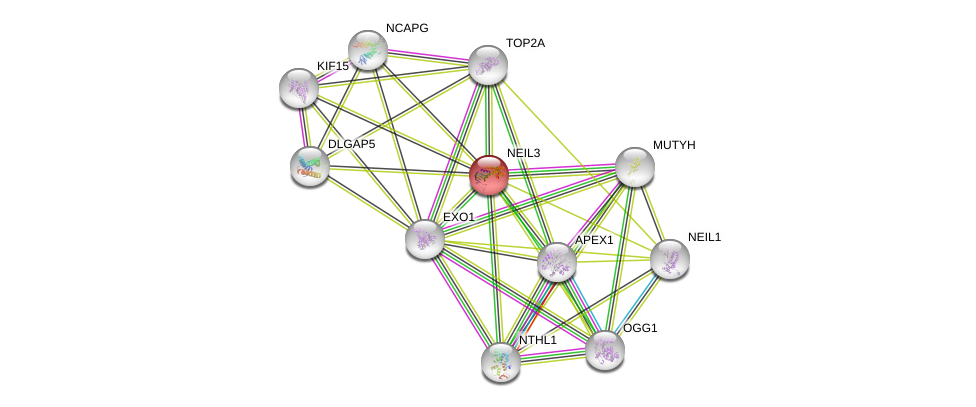
| hsa03410 | Base excision repair | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000109674 | Subcutaneous Fat | 2.45388273298897E-5 | 17903300 |
| ENSG00000109674 | Subcutaneous Fat | 2.87687391937741E-5 | 17903300 |
| ENSG00000109674 | Abdominal Fat | 1.93592959196742E-6 | 17903300 |
| ENSG00000109674 | Abdominal Fat | 2.13718218899928E-6 | 17903300 |
| ENSG00000109674 | Intra-Abdominal Fat | 3.05938989166865E-5 | 17903300 |
| ENSG00000109674 | Body Weights and Measures | 3.53412189277869E-5 | 17903300 |
| ENSG00000109674 | Heart Rate | 9.1026905905563E-7 | 17903306 |
| ENSG00000109674 | Heart Rate | 6.88670489997163E-6 | 17903306 |
| ENSG00000109674 | Glucose | 9.356e-005 | - |
| ENSG00000109674 | Platelet Function Tests | 2.1314100E-005 | - |
| ENSG00000109674 | Platelet Function Tests | 2.1314100E-005 | - |
| ENSG00000109674 | Selenium | 3E-7 | 23698163 |
| ENSG00000109674 | Heart Rate | 7E-6 | 17903306 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000109674 | NEIL3 | 100 | 51.064 | ENSACIG00000002265 | neil3 | 100 | 51.064 | Amphilophus_citrinellus |
| ENSG00000109674 | NEIL3 | 100 | 51.774 | ENSATEG00000004193 | neil3 | 100 | 51.774 | Anabas_testudineus |
| ENSG00000109674 | NEIL3 | 100 | 84.488 | ENSANAG00000026464 | NEIL3 | 100 | 84.488 | Aotus_nancymaae |
| ENSG00000109674 | NEIL3 | 100 | 51.452 | ENSACLG00000011270 | neil3 | 100 | 52.389 | Astatotilapia_calliptera |
| ENSG00000109674 | NEIL3 | 100 | 78.418 | ENSBTAG00000005825 | NEIL3 | 100 | 78.418 | Bos_taurus |
| ENSG00000109674 | NEIL3 | 100 | 90.759 | ENSCJAG00000005464 | NEIL3 | 100 | 90.759 | Callithrix_jacchus |
| ENSG00000109674 | NEIL3 | 100 | 78.089 | ENSCHIG00000009157 | NEIL3 | 100 | 78.089 | Capra_hircus |
| ENSG00000109674 | NEIL3 | 100 | 81.188 | ENSTSYG00000014680 | NEIL3 | 100 | 81.188 | Carlito_syrichta |
| ENSG00000109674 | NEIL3 | 85 | 77.973 | ENSCAPG00000006333 | NEIL3 | 98 | 77.973 | Cavia_aperea |
| ENSG00000109674 | NEIL3 | 100 | 78.254 | ENSCPOG00000003297 | NEIL3 | 100 | 78.254 | Cavia_porcellus |
| ENSG00000109674 | NEIL3 | 100 | 91.419 | ENSCCAG00000018838 | NEIL3 | 100 | 91.419 | Cebus_capucinus |
| ENSG00000109674 | NEIL3 | 100 | 77.265 | ENSCLAG00000015661 | NEIL3 | 100 | 77.265 | Chinchilla_lanigera |
| ENSG00000109674 | NEIL3 | 100 | 90.083 | ENSCSAG00000003064 | NEIL3 | 100 | 90.083 | Chlorocebus_sabaeus |
| ENSG00000109674 | NEIL3 | 77 | 89.831 | ENSCHOG00000008101 | NEIL3 | 77 | 89.831 | Choloepus_hoffmanni |
| ENSG00000109674 | NEIL3 | 100 | 91.074 | ENSCANG00000043622 | NEIL3 | 100 | 91.074 | Colobus_angolensis_palliatus |
| ENSG00000109674 | NEIL3 | 100 | 52.335 | ENSCSEG00000003828 | neil3 | 99 | 52.335 | Cynoglossus_semilaevis |
| ENSG00000109674 | NEIL3 | 100 | 51.060 | ENSDARG00000020079 | neil3 | 100 | 51.060 | Danio_rerio |
| ENSG00000109674 | NEIL3 | 86 | 80.614 | ENSDNOG00000012236 | NEIL3 | 100 | 80.614 | Dasypus_novemcinctus |
| ENSG00000109674 | NEIL3 | 100 | 80.395 | ENSEASG00005022272 | NEIL3 | 100 | 80.395 | Equus_asinus_asinus |
| ENSG00000109674 | NEIL3 | 100 | 79.538 | ENSECAG00000009121 | NEIL3 | 100 | 79.901 | Equus_caballus |
| ENSG00000109674 | NEIL3 | 75 | 70.485 | ENSEEUG00000010848 | NEIL3 | 100 | 70.485 | Erinaceus_europaeus |
| ENSG00000109674 | NEIL3 | 100 | 78.089 | ENSFCAG00000004956 | NEIL3 | 100 | 78.089 | Felis_catus |
| ENSG00000109674 | NEIL3 | 100 | 50.647 | ENSHBUG00000023848 | neil3 | 100 | 50.647 | Haplochromis_burtoni |
| ENSG00000109674 | NEIL3 | 86 | 79.271 | ENSHGLG00000006643 | NEIL3 | 100 | 79.271 | Heterocephalus_glaber_female |
| ENSG00000109674 | NEIL3 | 86 | 79.271 | ENSHGLG00100006959 | NEIL3 | 100 | 79.271 | Heterocephalus_glaber_male |
| ENSG00000109674 | NEIL3 | 100 | 80.066 | ENSSTOG00000016245 | NEIL3 | 100 | 80.066 | Ictidomys_tridecemlineatus |
| ENSG00000109674 | NEIL3 | 63 | 56.701 | ENSLACG00000000308 | NEIL3 | 99 | 56.701 | Latimeria_chalumnae |
| ENSG00000109674 | NEIL3 | 100 | 92.066 | ENSMFAG00000002547 | NEIL3 | 100 | 92.066 | Macaca_fascicularis |
| ENSG00000109674 | NEIL3 | 100 | 91.901 | ENSMMUG00000007394 | NEIL3 | 100 | 91.901 | Macaca_mulatta |
| ENSG00000109674 | NEIL3 | 100 | 91.901 | ENSMNEG00000036177 | NEIL3 | 100 | 91.901 | Macaca_nemestrina |
| ENSG00000109674 | NEIL3 | 100 | 51.967 | ENSMAMG00000017052 | neil3 | 100 | 51.864 | Mastacembelus_armatus |
| ENSG00000109674 | NEIL3 | 100 | 51.613 | ENSMZEG00005001680 | neil3 | 100 | 51.613 | Maylandia_zebra |
| ENSG00000109674 | NEIL3 | 90 | 63.815 | ENSMODG00000003813 | NEIL3 | 99 | 63.815 | Monodelphis_domestica |
| ENSG00000109674 | NEIL3 | 100 | 74.092 | MGP_CAROLIEiJ_G0031052 | Neil3 | 100 | 74.092 | Mus_caroli |
| ENSG00000109674 | NEIL3 | 100 | 73.927 | ENSMUSG00000039396 | Neil3 | 100 | 73.927 | Mus_musculus |
| ENSG00000109674 | NEIL3 | 100 | 73.762 | MGP_PahariEiJ_G0021518 | Neil3 | 100 | 73.762 | Mus_pahari |
| ENSG00000109674 | NEIL3 | 100 | 74.257 | MGP_SPRETEiJ_G0032167 | Neil3 | 100 | 74.257 | Mus_spretus |
| ENSG00000109674 | NEIL3 | 100 | 50.330 | ENSNBRG00000013229 | neil3 | 100 | 50.330 | Neolamprologus_brichardi |
| ENSG00000109674 | NEIL3 | 100 | 94.554 | ENSNLEG00000016818 | NEIL3 | 100 | 94.554 | Nomascus_leucogenys |
| ENSG00000109674 | NEIL3 | 100 | 76.606 | ENSODEG00000004102 | NEIL3 | 100 | 76.606 | Octodon_degus |
| ENSG00000109674 | NEIL3 | 100 | 51.133 | ENSONIG00000001176 | neil3 | 100 | 51.133 | Oreochromis_niloticus |
| ENSG00000109674 | NEIL3 | 99 | 80.167 | ENSOCUG00000001040 | NEIL3 | 99 | 80.167 | Oryctolagus_cuniculus |
| ENSG00000109674 | NEIL3 | 100 | 48.544 | ENSOMEG00000014798 | neil3 | 100 | 48.544 | Oryzias_melastigma |
| ENSG00000109674 | NEIL3 | 100 | 81.219 | ENSOGAG00000012624 | NEIL3 | 100 | 81.219 | Otolemur_garnettii |
| ENSG00000109674 | NEIL3 | 96 | 76.936 | ENSOARG00000006852 | NEIL3 | 100 | 79.459 | Ovis_aries |
| ENSG00000109674 | NEIL3 | 100 | 96.370 | ENSPPAG00000035318 | NEIL3 | 100 | 96.370 | Pan_paniscus |
| ENSG00000109674 | NEIL3 | 100 | 78.418 | ENSPPRG00000003958 | NEIL3 | 100 | 78.418 | Panthera_pardus |
| ENSG00000109674 | NEIL3 | 100 | 78.913 | ENSPTIG00000012966 | NEIL3 | 100 | 78.913 | Panthera_tigris_altaica |
| ENSG00000109674 | NEIL3 | 100 | 97.525 | ENSPTRG00000016617 | NEIL3 | 100 | 97.525 | Pan_troglodytes |
| ENSG00000109674 | NEIL3 | 100 | 91.901 | ENSPANG00000014727 | NEIL3 | 100 | 91.901 | Papio_anubis |
| ENSG00000109674 | NEIL3 | 100 | 50.243 | ENSPFOG00000018858 | neil3 | 100 | 50.243 | Poecilia_formosa |
| ENSG00000109674 | NEIL3 | 100 | 49.919 | ENSPLAG00000010685 | neil3 | 100 | 49.919 | Poecilia_latipinna |
| ENSG00000109674 | NEIL3 | 100 | 50.243 | ENSPMEG00000013971 | neil3 | 100 | 50.243 | Poecilia_mexicana |
| ENSG00000109674 | NEIL3 | 100 | 50.247 | ENSPREG00000009841 | neil3 | 100 | 50.247 | Poecilia_reticulata |
| ENSG00000109674 | NEIL3 | 68 | 83.051 | ENSPCAG00000006386 | NEIL3 | 81 | 83.051 | Procavia_capensis |
| ENSG00000109674 | NEIL3 | 77 | 90.756 | ENSPCOG00000010888 | NEIL3 | 97 | 90.756 | Propithecus_coquereli |
| ENSG00000109674 | NEIL3 | 99 | 77.483 | ENSPVAG00000002255 | NEIL3 | 99 | 77.483 | Pteropus_vampyrus |
| ENSG00000109674 | NEIL3 | 100 | 51.303 | ENSPNYG00000021276 | neil3 | 100 | 51.303 | Pundamilia_nyererei |
| ENSG00000109674 | NEIL3 | 98 | 47.892 | ENSPNAG00000019983 | neil3 | 100 | 47.892 | Pygocentrus_nattereri |
| ENSG00000109674 | NEIL3 | 100 | 72.607 | ENSRNOG00000011688 | Neil3 | 100 | 72.607 | Rattus_norvegicus |
| ENSG00000109674 | NEIL3 | 100 | 90.413 | ENSRBIG00000023002 | NEIL3 | 100 | 90.413 | Rhinopithecus_bieti |
| ENSG00000109674 | NEIL3 | 100 | 90.248 | ENSRROG00000000069 | NEIL3 | 100 | 90.248 | Rhinopithecus_roxellana |
| ENSG00000109674 | NEIL3 | 100 | 90.759 | ENSSBOG00000032601 | NEIL3 | 100 | 90.759 | Saimiri_boliviensis_boliviensis |
| ENSG00000109674 | NEIL3 | 100 | 67.105 | ENSSHAG00000007680 | NEIL3 | 100 | 67.105 | Sarcophilus_harrisii |
| ENSG00000109674 | NEIL3 | 100 | 51.442 | ENSSMAG00000009621 | neil3 | 100 | 51.763 | Scophthalmus_maximus |
| ENSG00000109674 | NEIL3 | 100 | 78.418 | ENSSSCG00000015771 | NEIL3 | 100 | 78.418 | Sus_scrofa |
| ENSG00000109674 | NEIL3 | 100 | 78.254 | ENSTTRG00000013224 | NEIL3 | 100 | 78.254 | Tursiops_truncatus |
| ENSG00000109674 | NEIL3 | 100 | 69.737 | ENSVPAG00000005020 | NEIL3 | 100 | 69.737 | Vicugna_pacos |
| ENSG00000109674 | NEIL3 | 90 | 48.087 | ENSXCOG00000014831 | neil3 | 99 | 48.087 | Xiphophorus_couchianus |
| ENSG00000109674 | NEIL3 | 100 | 49.838 | ENSXMAG00000028638 | neil3 | 100 | 49.838 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000405 | bubble DNA binding | - | ISS | Function |
| GO:0003684 | damaged DNA binding | - | IEA | Function |
| GO:0003690 | double-stranded DNA binding | - | ISS | Function |
| GO:0003697 | single-stranded DNA binding | 19170771. | IDA | Function |
| GO:0003906 | DNA-(apurinic or apyrimidinic site) endonuclease activity | 21873635. | IBA | Function |
| GO:0003906 | DNA-(apurinic or apyrimidinic site) endonuclease activity | 19170771. | IDA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 12433996. | IDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0006284 | base-excision repair | 21873635. | IBA | Process |
| GO:0006284 | base-excision repair | 23755964. | IDA | Process |
| GO:0006284 | base-excision repair | - | ISS | Process |
| GO:0006289 | nucleotide-excision repair | - | IEA | Process |
| GO:0008270 | zinc ion binding | - | IEA | Function |
| GO:0019104 | DNA N-glycosylase activity | 21873635. | IBA | Function |
| GO:0019104 | DNA N-glycosylase activity | 23755964. | IDA | Function |
| GO:0019104 | DNA N-glycosylase activity | - | ISS | Function |
| GO:0140078 | class I DNA-(apurinic or apyrimidinic site) endonuclease activity | - | IEA | Function |