
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| LIHC | |||
| READ |
| Cancer | P-value | Q-value |
|---|---|---|
| ACC | ||
| SKCM | ||
| BRCA | ||
| KIRP | ||
| PAAD | ||
| LAML | ||
| LIHC | ||
| LUAD | ||
| UVM |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 23137539 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000514106 | UGDH-210 | 568 | - | ENSP00000425834 | 153 (aa) | - | E7ETF4 |
| ENST00000316423 | UGDH-201 | 3216 | XM_005262667 | ENSP00000319501 | 494 (aa) | XP_005262724 | O60701 |
| ENST00000501493 | UGDH-202 | 2826 | - | ENSP00000422909 | 427 (aa) | - | O60701 |
| ENST00000510881 | UGDH-209 | 549 | - | ENSP00000424059 | 55 (aa) | - | E9PBD2 |
| ENST00000515398 | UGDH-212 | 584 | - | - | - (aa) | - | - |
| ENST00000503779 | UGDH-203 | 575 | - | ENSP00000424435 | 55 (aa) | - | E9PBD2 |
| ENST00000507089 | UGDH-206 | 1712 | - | ENSP00000426560 | 397 (aa) | - | O60701 |
| ENST00000515021 | UGDH-211 | 861 | - | ENSP00000421954 | 181 (aa) | - | E7EV97 |
| ENST00000505698 | UGDH-204 | 433 | - | ENSP00000422565 | 84 (aa) | - | E7ER95 |
| ENST00000510490 | UGDH-208 | 534 | - | ENSP00000427708 | 53 (aa) | - | D6RHF4 |
| ENST00000509391 | UGDH-207 | 577 | - | ENSP00000422603 | 130 (aa) | - | E7ER83 |
| ENST00000506179 | UGDH-205 | 3021 | - | ENSP00000421757 | 494 (aa) | - | O60701 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000109814 | Platelet Function Tests | 2.9673700E-005 | - |
| ENSG00000109814 | Platelet Function Tests | 2.9673700E-005 | - |
| ENSG00000109814 | Platelet Function Tests | 2.8331100E-005 | - |
| ENSG00000109814 | Platelet Function Tests | 2.1238000E-005 | - |
| ENSG00000109814 | Platelet Function Tests | 3.5690800E-005 | - |
| ENSG00000109814 | Platelet Function Tests | 3.5690800E-005 | - |
| ENSG00000109814 | Platelet Function Tests | 3.3300400E-005 | - |
| ENSG00000109814 | Platelet Function Tests | 2.8975700E-005 | - |
| ENSG00000109814 | Autistic Disorder | 9.0510000E-006 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000109814 | rs56118664 | 4 | 39516245 | G | Ankle injury | 28957384 | 1.8994 | EFO_1002021 | |
| ENSG00000109814 | rs3755899 | 4 | 39528080 | G | Peripheral arterial disease (traffic-related air pollution interaction) | 27082954 | [1772.72-1806.94] | 1789.83 | EFO_0004265|EFO_0007908 |
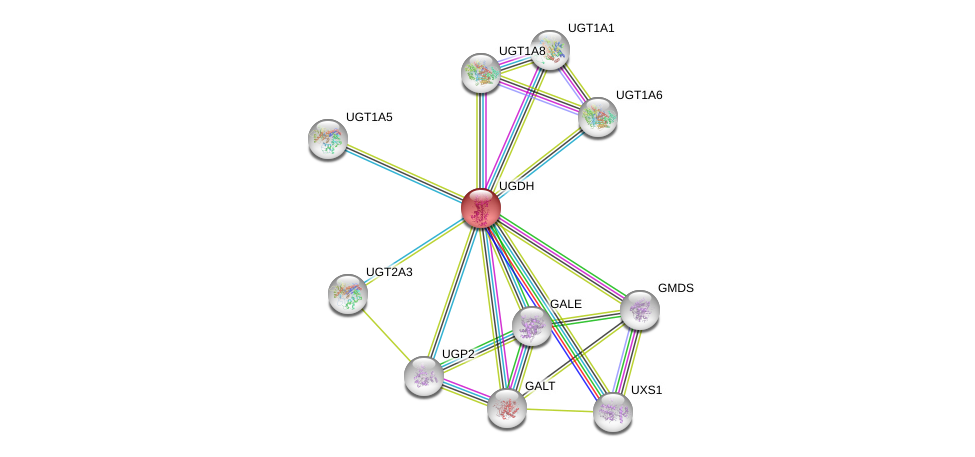
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000109814 | UGDH | 98 | 64.062 | WBGene00005022 | sqv-4 | 95 | 65.659 | Caenorhabditis_elegans |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001702 | gastrulation with mouth forming second | - | ISS | Process |
| GO:0003979 | UDP-glucose 6-dehydrogenase activity | 27966912. | IDA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005975 | carbohydrate metabolic process | - | IEA | Process |
| GO:0006024 | glycosaminoglycan biosynthetic process | 21873635. | IBA | Process |
| GO:0006065 | UDP-glucuronate biosynthetic process | 27966912. | IDA | Process |
| GO:0006065 | UDP-glucuronate biosynthetic process | - | IEA | Process |
| GO:0006065 | UDP-glucuronate biosynthetic process | - | TAS | Process |
| GO:0015012 | heparan sulfate proteoglycan biosynthetic process | - | ISS | Process |
| GO:0030206 | chondroitin sulfate biosynthetic process | - | ISS | Process |
| GO:0034214 | protein hexamerization | 27966912. | IDA | Process |
| GO:0051287 | NAD binding | - | IEA | Function |
| GO:0055114 | oxidation-reduction process | - | IEA | Process |
| GO:0070062 | extracellular exosome | 23533145. | HDA | Component |