
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BLCA | |||
| CESC | |||
| CHOL | |||
| ESCA | |||
| KIRP | |||
| LGG | |||
| LIHC | |||
| LUAD | |||
| OV | |||
| READ | |||
| SARC | |||
| STAD | |||
| TGCT | |||
| UCEC | |||
| UCS | |||
| UVM |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| BRCA | ||
| COAD | ||
| CESC | ||
| READ | ||
| LAML | ||
| KICH | ||
| LIHC |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000526686 | HSPA8-208 | 740 | - | ENSP00000435019 | 150 (aa) | - | E9PM13 |
| ENST00000532167 | HSPA8-216 | 570 | - | - | - (aa) | - | - |
| ENST00000524552 | HSPA8-203 | 963 | - | ENSP00000435908 | 223 (aa) | - | E9PS65 |
| ENST00000526862 | HSPA8-209 | 504 | - | - | - (aa) | - | - |
| ENST00000527983 | HSPA8-211 | 1509 | - | - | - (aa) | - | - |
| ENST00000532636 | HSPA8-218 | 2306 | - | ENSP00000437125 | 646 (aa) | - | P11142 |
| ENST00000534624 | HSPA8-224 | 2463 | - | ENSP00000432083 | 646 (aa) | - | P11142 |
| ENST00000530391 | HSPA8-213 | 648 | - | ENSP00000434851 | 132 (aa) | - | E9PN25 |
| ENST00000227378 | HSPA8-201 | 2186 | XM_011542798 | ENSP00000227378 | 646 (aa) | XP_011541100 | P11142 |
| ENST00000532091 | HSPA8-215 | 2301 | - | - | - (aa) | - | - |
| ENST00000525463 | HSPA8-205 | 583 | - | ENSP00000436762 | 168 (aa) | - | E9PI65 |
| ENST00000533238 | HSPA8-220 | 465 | - | - | - (aa) | - | - |
| ENST00000453788 | HSPA8-202 | 2004 | - | ENSP00000404372 | 493 (aa) | - | P11142 |
| ENST00000532780 | HSPA8-219 | 1194 | - | - | - (aa) | - | - |
| ENST00000534319 | HSPA8-222 | 1884 | - | ENSP00000433316 | 410 (aa) | - | A8K7Q2 |
| ENST00000526110 | HSPA8-207 | 1924 | - | ENSP00000433584 | 627 (aa) | - | E9PKE3 |
| ENST00000525624 | HSPA8-206 | 1048 | - | ENSP00000435154 | 187 (aa) | - | E9PLF4 |
| ENST00000532182 | HSPA8-217 | 674 | - | ENSP00000434415 | 171 (aa) | - | E9PQQ4 |
| ENST00000534567 | HSPA8-223 | 588 | - | ENSP00000431641 | 183 (aa) | - | E9PK54 |
| ENST00000531063 | HSPA8-214 | 578 | - | - | - (aa) | - | - |
| ENST00000524590 | HSPA8-204 | 536 | - | ENSP00000434565 | 137 (aa) | - | E9PPY6 |
| ENST00000527387 | HSPA8-210 | 579 | - | ENSP00000436183 | 178 (aa) | - | E9PQK7 |
| ENST00000528292 | HSPA8-212 | 1112 | - | ENSP00000432884 | 312 (aa) | - | E9PN89 |
| ENST00000533540 | HSPA8-221 | 1826 | - | ENSP00000437189 | 500 (aa) | - | E9PNE6 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa03040 | Spliceosome | KEGG |
| hsa04010 | MAPK signaling pathway | KEGG |
| hsa04141 | Protein processing in endoplasmic reticulum | KEGG |
| hsa04144 | Endocytosis | KEGG |
| hsa04213 | Longevity regulating pathway - multiple species | KEGG |
| hsa04612 | Antigen processing and presentation | KEGG |
| hsa04915 | Estrogen signaling pathway | KEGG |
| hsa05134 | Legionellosis | KEGG |
| hsa05145 | Toxoplasmosis | KEGG |
| hsa05162 | Measles | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000109971 | rs4936770 | 11 | 123058167 | ? | Time-dependent creatinine clearance change response to tenofovir treatment in HIV infection (time and treatment arm interaction) | 26148204 | EFO_0009279|EFO_0000764|EFO_0007934 | ||
| ENSG00000109971 | rs4802 | 11 | 123057914 | ? | Time-dependent creatinine clearance change response to tenofovir treatment in HIV infection (time and treatment arm interaction) | 26148204 | EFO_0009279|EFO_0000764|EFO_0007934 | ||
| ENSG00000109971 | rs4936770 | 11 | 123058167 | ? | Time-dependent creatinine clearance change response to tenofovir treatment in HIV infection (time and treatment arm interaction) | 26148204 | EFO_0009279|EFO_0000764|EFO_0007934 | ||
| ENSG00000109971 | rs2236659 | 11 | 123062473 | ? | Mean corpuscular hemoglobin | 30595370 | EFO_0004527 |
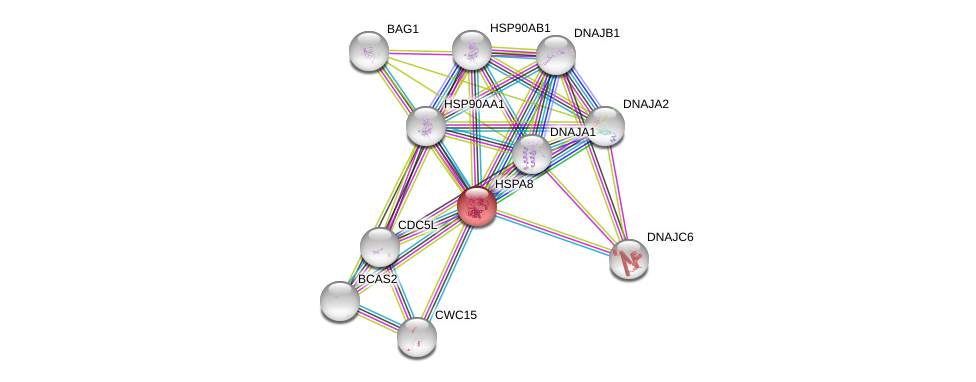
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000109971 | HSPA8 | 100 | 100.000 | ENSMUSG00000015656 | Hspa8 | 100 | 100.000 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000151 | ubiquitin ligase complex | 12150907. | IDA | Component |
| GO:0000398 | mRNA splicing, via spliceosome | - | TAS | Process |
| GO:0000974 | Prp19 complex | 20176811. | IDA | Component |
| GO:0001664 | G protein-coupled receptor binding | 12150907. | IPI | Function |
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0005515 | protein binding | 9305631.9499401.10722728.10954706.14532270.14557246.15047060.15657067.15708368.15936278.16169070.16189514.16275660.16293251.16531398.17022977.17400507.17620599.18457437.19338310.19800331.20195357.20391533.20618441.21148293.21150129.22085931.22365833.22645275.22990239.23431407.23455607.23523103.23801752.24122553.24136289.24189400.24318877.24510904.24658140.24787902.24947832.25416956.25609649.25719862.25760597.25910212.26212789.26871637.27103069.27708256.27916661.29568061.29601588.29764935.30021884. | IPI | Function |
| GO:0005524 | ATP binding | 21873635. | IBA | Function |
| GO:0005524 | ATP binding | 23921388. | IDA | Function |
| GO:0005576 | extracellular region | - | TAS | Component |
| GO:0005615 | extracellular space | 16502470. | HDA | Component |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 20176811. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005681 | spliceosomal complex | - | IEA | Component |
| GO:0005730 | nucleolus | - | IEA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005764 | lysosome | 21873635. | IBA | Component |
| GO:0005765 | lysosomal membrane | - | ISS | Component |
| GO:0005776 | autophagosome | 21873635. | IBA | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | 21231916. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005886 | plasma membrane | 21873635. | IBA | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0005925 | focal adhesion | 21423176. | HDA | Component |
| GO:0006457 | protein folding | 8530083. | NAS | Process |
| GO:0006986 | response to unfolded protein | 21873635. | IBA | Process |
| GO:0006986 | response to unfolded protein | 11093761. | NAS | Process |
| GO:0007269 | neurotransmitter secretion | - | TAS | Process |
| GO:0008088 | axo-dendritic transport | 21873635. | IBA | Process |
| GO:0009267 | cellular response to starvation | 20176123. | TAS | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0016192 | vesicle-mediated transport | 21873635. | IBA | Process |
| GO:0016887 | ATPase activity | 21873635. | IBA | Function |
| GO:0016887 | ATPase activity | 23921388. | IDA | Function |
| GO:0016887 | ATPase activity | - | TAS | Function |
| GO:0019221 | cytokine-mediated signaling pathway | - | TAS | Process |
| GO:0019899 | enzyme binding | 23921388. | IPI | Function |
| GO:0023026 | MHC class II protein complex binding | 20458337. | HDA | Function |
| GO:0030424 | axon | 21873635. | IBA | Component |
| GO:0030425 | dendrite | 21873635. | IBA | Component |
| GO:0030674 | protein binding, bridging | 16207813. | IPI | Function |
| GO:0031072 | heat shock protein binding | 21873635. | IBA | Function |
| GO:0031072 | heat shock protein binding | 17182002.23921388. | IPI | Function |
| GO:0031625 | ubiquitin protein ligase binding | 12150907.16207813. | IPI | Function |
| GO:0031647 | regulation of protein stability | 26212789. | IMP | Process |
| GO:0034605 | cellular response to heat | 21873635. | IBA | Process |
| GO:0034620 | cellular response to unfolded protein | 21873635. | IBA | Process |
| GO:0034774 | secretory granule lumen | - | TAS | Component |
| GO:0042026 | protein refolding | 21873635. | IBA | Process |
| GO:0042026 | protein refolding | 21231916.25719862. | IDA | Process |
| GO:0042470 | melanosome | - | IEA | Component |
| GO:0042623 | ATPase activity, coupled | 21873635. | IBA | Function |
| GO:0042623 | ATPase activity, coupled | 8530083. | NAS | Function |
| GO:0042623 | ATPase activity, coupled | - | TAS | Function |
| GO:0043195 | terminal bouton | 21873635. | IBA | Component |
| GO:0043202 | lysosomal lumen | 25719862. | TAS | Component |
| GO:0043254 | regulation of protein complex assembly | 20176123. | TAS | Process |
| GO:0043312 | neutrophil degranulation | - | TAS | Process |
| GO:0043488 | regulation of mRNA stability | - | TAS | Process |
| GO:0044183 | protein binding involved in protein folding | 21873635. | IBA | Function |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0045892 | negative regulation of transcription, DNA-templated | 10722728. | IDA | Process |
| GO:0046034 | ATP metabolic process | 23921388. | IDA | Process |
| GO:0051082 | unfolded protein binding | 21873635. | IBA | Function |
| GO:0051082 | unfolded protein binding | 21231916. | IDA | Function |
| GO:0051085 | chaperone cofactor-dependent protein refolding | 21873635. | IBA | Process |
| GO:0051087 | chaperone binding | 16207813. | IPI | Function |
| GO:0051787 | misfolded protein binding | 21873635. | IBA | Function |
| GO:0055131 | C3HC4-type RING finger domain binding | 25281747. | IPI | Function |
| GO:0061024 | membrane organization | - | TAS | Process |
| GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane | - | TAS | Component |
| GO:0061635 | regulation of protein complex stability | - | ISS | Process |
| GO:0061684 | chaperone-mediated autophagy | - | ISS | Process |
| GO:0061684 | chaperone-mediated autophagy | 20176123.25719862. | TAS | Process |
| GO:0061738 | late endosomal microautophagy | 21873635. | IBA | Process |
| GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy | 21873635. | IBA | Process |
| GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy | 26212789. | IMP | Process |
| GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy | 2799391. | TAS | Process |
| GO:0061741 | chaperone-mediated protein transport involved in chaperone-mediated autophagy | 21873635. | IBA | Process |
| GO:0061741 | chaperone-mediated protein transport involved in chaperone-mediated autophagy | 25719862. | NAS | Process |
| GO:0070062 | extracellular exosome | 19056867.19199708.20458337.21362503.23533145. | HDA | Component |
| GO:0070062 | extracellular exosome | 19028452.21235781. | IDA | Component |
| GO:0072318 | clathrin coat disassembly | - | IEA | Process |
| GO:0072562 | blood microparticle | 22516433. | HDA | Component |
| GO:0098575 | lumenal side of lysosomal membrane | 20176123. | TAS | Component |
| GO:0099523 | presynaptic cytosol | 21873635. | IBA | Component |
| GO:0099524 | postsynaptic cytosol | 21873635. | IBA | Component |
| GO:0101031 | chaperone complex | 16207813. | IPI | Component |
| GO:1900034 | regulation of cellular response to heat | - | TAS | Process |
| GO:1902904 | negative regulation of supramolecular fiber organization | 23921388. | IDA | Process |
| GO:1904589 | regulation of protein import | 20176123. | TAS | Process |
| GO:1904764 | chaperone-mediated autophagy translocation complex disassembly | 21873635. | IBA | Process |
| GO:1904764 | chaperone-mediated autophagy translocation complex disassembly | - | ISS | Process |
| GO:1904813 | ficolin-1-rich granule lumen | - | TAS | Component |
| GO:1990832 | slow axonal transport | 21873635. | IBA | Process |
| GO:1990833 | clathrin-uncoating ATPase activity | 21873635. | IBA | Function |
| GO:1990904 | ribonucleoprotein complex | 17289661. | IDA | Component |