
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PCPG | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| LAML | |||
| LUAD | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| STAD | |||
| THCA | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| MESO | ||
| ACC | ||
| UCS | ||
| ESCA | ||
| COAD | ||
| READ | ||
| LAML | ||
| KICH | ||
| LIHC | ||
| LGG | ||
| UVM | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000504197 | H2AFY-206 | 498 | - | - | - (aa) | - | - |
| ENST00000304332 | H2AFY-201 | 1881 | XM_011543730 | ENSP00000302572 | 371 (aa) | XP_011542032 | O75367 |
| ENST00000511689 | H2AFY-217 | 2789 | XM_011543731 | ENSP00000423563 | 372 (aa) | XP_011542033 | O75367 |
| ENST00000451949 | H2AFY-205 | 1744 | - | - | - (aa) | - | - |
| ENST00000510038 | H2AFY-215 | 1314 | XM_011543729 | ENSP00000424971 | 372 (aa) | XP_011542031 | O75367 |
| ENST00000512507 | H2AFY-218 | 10982 | - | - | - (aa) | - | - |
| ENST00000423969 | H2AFY-204 | 924 | - | ENSP00000415121 | 200 (aa) | - | B4DJC3 |
| ENST00000506671 | H2AFY-210 | 1905 | - | ENSP00000423718 | 165 (aa) | - | D6RCF2 |
| ENST00000312469 | H2AFY-202 | 1859 | XM_005272132 | ENSP00000310169 | 369 (aa) | XP_005272189 | O75367 |
| ENST00000513210 | H2AFY-219 | 775 | - | - | - (aa) | - | - |
| ENST00000360597 | H2AFY-203 | 1772 | - | - | - (aa) | - | - |
| ENST00000506532 | H2AFY-209 | 677 | - | - | - (aa) | - | - |
| ENST00000508120 | H2AFY-212 | 536 | - | - | - (aa) | - | - |
| ENST00000513268 | H2AFY-220 | 501 | - | - | - (aa) | - | - |
| ENST00000511494 | H2AFY-216 | 2521 | - | - | - (aa) | - | - |
| ENST00000508785 | H2AFY-213 | 966 | - | - | - (aa) | - | - |
| ENST00000507868 | H2AFY-211 | 861 | - | - | - (aa) | - | - |
| ENST00000506218 | H2AFY-208 | 758 | - | - | - (aa) | - | - |
| ENST00000505827 | H2AFY-207 | 2769 | - | - | - (aa) | - | - |
| ENST00000508962 | H2AFY-214 | 293 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000113648 | rs79237520 | 5 | 135376876 | T | Lymphocyte counts | 27863252 | [0.063-0.111] unit increase | 0.08702981 | EFO_0004587 |
| ENSG00000113648 | rs703254 | 5 | 135394553 | ? | Adolescent idiopathic scoliosis | 30019117 | EFO_0005423 |
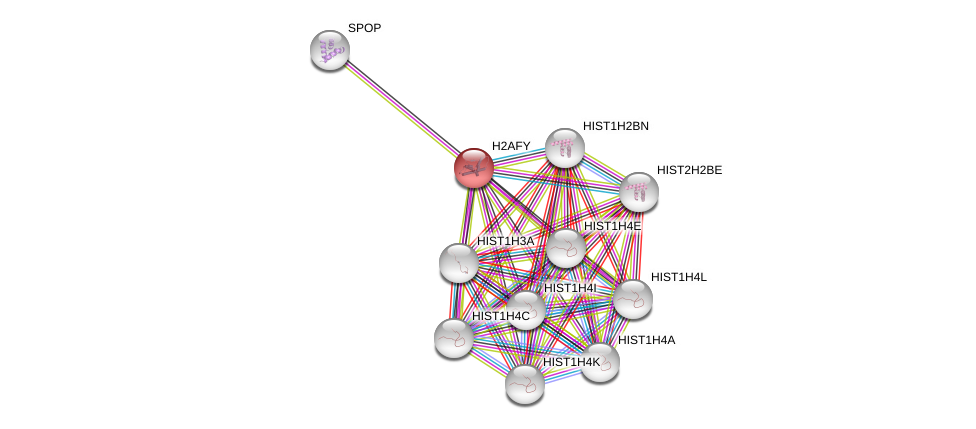
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000113648 | H2AFY | 100 | 98.387 | ENSMUSG00000015937 | H2afy | 100 | 98.387 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000122 | negative regulation of transcription by RNA polymerase II | 19734898. | IMP | Process |
| GO:0000182 | rDNA binding | 24071584. | IDA | Function |
| GO:0000228 | nuclear chromosome | 18936163. | IDA | Component |
| GO:0000784 | nuclear chromosome, telomeric region | 19135898. | HDA | Component |
| GO:0000786 | nucleosome | 9714746. | NAS | Component |
| GO:0000790 | nuclear chromatin | 21873635. | IBA | Component |
| GO:0000790 | nuclear chromatin | 19734898.22194607.24071584. | IDA | Component |
| GO:0000793 | condensed chromosome | - | IEA | Component |
| GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | 23595991. | IDA | Function |
| GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding | 25526730. | IDA | Function |
| GO:0001739 | sex chromatin | 19486527. | TAS | Component |
| GO:0001740 | Barr body | 11331621.18936163. | IDA | Component |
| GO:0003677 | DNA binding | 21873635. | IBA | Function |
| GO:0003677 | DNA binding | 9714746. | NAS | Function |
| GO:0005515 | protein binding | 22391447.22589551.25416956.25609649. | IPI | Function |
| GO:0005634 | nucleus | 21630459. | HDA | Component |
| GO:0005634 | nucleus | - | IDA | Component |
| GO:0005721 | pericentric heterochromatin | 19486527. | IDA | Component |
| GO:0005730 | nucleolus | 24071584. | IDA | Component |
| GO:0006325 | chromatin organization | 21873635. | IBA | Process |
| GO:0006334 | nucleosome assembly | 9714746. | NAS | Process |
| GO:0007549 | dosage compensation | 15053874. | IDA | Process |
| GO:0010385 | double-stranded methylated DNA binding | 24071584. | IDA | Function |
| GO:0019216 | regulation of lipid metabolic process | 25526730. | IMP | Process |
| GO:0019899 | enzyme binding | 26373281. | IPI | Function |
| GO:0019901 | protein kinase binding | 22194607. | IPI | Function |
| GO:0030291 | protein serine/threonine kinase inhibitor activity | 22194607. | IMP | Function |
| GO:0031490 | chromatin DNA binding | 19734898.24071584. | IDA | Function |
| GO:0031492 | nucleosomal DNA binding | 20008927. | IDA | Function |
| GO:0031492 | nucleosomal DNA binding | - | ISS | Function |
| GO:0033128 | negative regulation of histone phosphorylation | 22194607. | IMP | Process |
| GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion | 26373281. | IDA | Process |
| GO:0040029 | regulation of gene expression, epigenetic | 20008927. | IMP | Process |
| GO:0044212 | transcription regulatory region DNA binding | 19734898.20008927. | IDA | Function |
| GO:0045618 | positive regulation of keratinocyte differentiation | 23595991. | IMP | Process |
| GO:0045814 | negative regulation of gene expression, epigenetic | 19734898.23595991.24071584. | IMP | Process |
| GO:0045815 | positive regulation of gene expression, epigenetic | 25526730. | IMP | Process |
| GO:0046982 | protein heterodimerization activity | - | IEA | Function |
| GO:0051572 | negative regulation of histone H3-K4 methylation | 25526730. | IMP | Process |
| GO:0061086 | negative regulation of histone H3-K27 methylation | 25526730. | IMP | Process |
| GO:0061187 | regulation of chromatin silencing at rDNA | 24071584. | IMP | Process |
| GO:0070062 | extracellular exosome | 23533145. | HDA | Component |
| GO:0071169 | establishment of protein localization to chromatin | 19734898. | IMP | Process |
| GO:0071901 | negative regulation of protein serine/threonine kinase activity | 22194607. | IMP | Process |
| GO:1901837 | negative regulation of transcription of nucleolar large rRNA by RNA polymerase I | 24071584. | IGI | Process |
| GO:1901837 | negative regulation of transcription of nucleolar large rRNA by RNA polymerase I | 24071584. | IMP | Process |
| GO:1902750 | negative regulation of cell cycle G2/M phase transition | 22194607. | IMP | Process |
| GO:1902882 | regulation of response to oxidative stress | 23022728. | IMP | Process |
| GO:1904815 | negative regulation of protein localization to chromosome, telomeric region | 26373281. | IDA | Process |
| GO:1990841 | promoter-specific chromatin binding | - | ISS | Function |