
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRC | |||
| KIRP | |||
| KIRP | |||
| LUAD | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| SKCM | |||
| THCA | |||
| UCEC |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| MESO | ||
| ACC | ||
| UCS | ||
| SKCM | ||
| BLCA | ||
| CESC | ||
| UCEC | ||
| LIHC | ||
| LGG |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000514833 | DBN1-211 | 456 | - | ENSP00000421465 | 124 (aa) | - | D6RFI1 |
| ENST00000292385 | DBN1-201 | 3072 | XM_011534446 | ENSP00000292385 | 651 (aa) | XP_011532748 | Q16643 |
| ENST00000309007 | DBN1-202 | 2995 | - | ENSP00000308532 | 649 (aa) | - | Q16643 |
| ENST00000505550 | DBN1-208 | 534 | - | - | - (aa) | - | - |
| ENST00000512501 | DBN1-210 | 1878 | - | ENSP00000423208 | 391 (aa) | - | D6R9Q9 |
| ENST00000393565 | DBN1-203 | 2701 | XM_005265827 | ENSP00000377195 | 695 (aa) | XP_005265884 | Q16643 |
| ENST00000472831 | DBN1-206 | 2667 | - | - | - (aa) | - | - |
| ENST00000467054 | DBN1-204 | 793 | - | - | - (aa) | - | - |
| ENST00000506117 | DBN1-209 | 477 | - | ENSP00000425546 | 93 (aa) | - | D6RCR4 |
| ENST00000477391 | DBN1-207 | 1044 | - | ENSP00000422854 | 317 (aa) | - | D6R9W4 |
| ENST00000471767 | DBN1-205 | 428 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000113758 | Heart Failure | 4.4110906E-005 | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000113758 | rs2545795 | 5 | 177460105 | A | Educational attainment (MTAG) | 30038396 | [0.01-0.016] unit increase | 0.013 | EFO_0004784 |
| ENSG00000113758 | rs2450333 | 5 | 177472545 | A | Intelligence | 29942086 | z-score decrease | 6.727 | EFO_0004337 |
| ENSG00000113758 | rs2545795 | 5 | 177460105 | A | Educational attainment (years of education) | 30038396 | [0.0098-0.0152] unit increase | 0.0125 | EFO_0004784 |
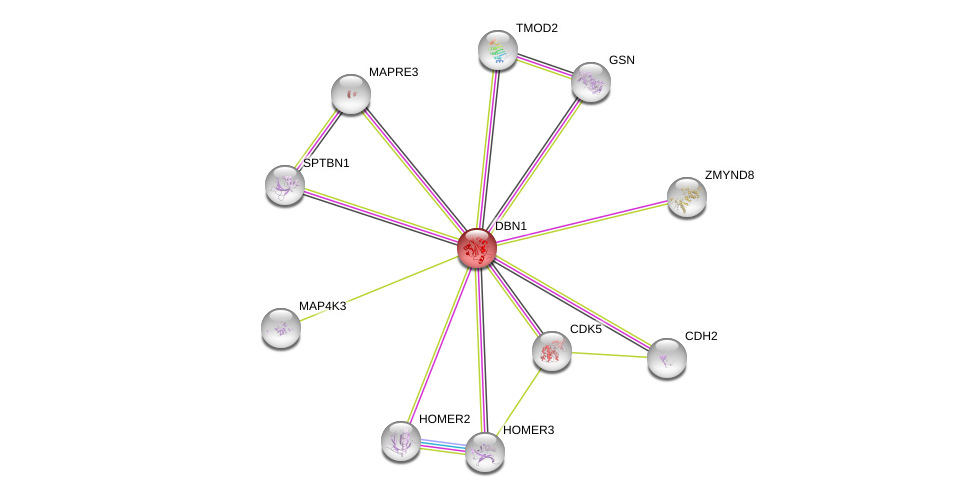
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003779 | actin binding | 12761245. | NAS | Function |
| GO:0005515 | protein binding | 12577067.20215400.21044950.23750010.23926103.23940795.25814554.25839164.28966017. | IPI | Function |
| GO:0005522 | profilin binding | - | ISS | Function |
| GO:0005737 | cytoplasm | 8838578.28966017. | IDA | Component |
| GO:0005737 | cytoplasm | 12009525. | NAS | Component |
| GO:0005856 | cytoskeleton | 24327345. | IDA | Component |
| GO:0005921 | gap junction | - | IEA | Component |
| GO:0007015 | actin filament organization | - | ISS | Process |
| GO:0010643 | cell communication by chemical coupling | - | IEA | Process |
| GO:0010644 | cell communication by electrical coupling | - | IEA | Process |
| GO:0014069 | postsynaptic density | - | IEA | Component |
| GO:0015629 | actin cytoskeleton | - | ISS | Component |
| GO:0030425 | dendrite | 8838578. | IDA | Component |
| GO:0030425 | dendrite | 12009525. | NAS | Component |
| GO:0030426 | growth cone | - | ISS | Component |
| GO:0030863 | cortical cytoskeleton | 17251419. | TAS | Component |
| GO:0031915 | positive regulation of synaptic plasticity | - | ISS | Process |
| GO:0032507 | maintenance of protein location in cell | - | IEA | Process |
| GO:0042641 | actomyosin | 12761245. | NAS | Component |
| GO:0045211 | postsynaptic membrane | - | IEA | Component |
| GO:0045296 | cadherin binding | 25468996. | HDA | Function |
| GO:0048168 | regulation of neuronal synaptic plasticity | 12009525. | NAS | Process |
| GO:0050773 | regulation of dendrite development | 12009525. | NAS | Process |
| GO:0051220 | cytoplasmic sequestering of protein | 28966017. | IDA | Process |
| GO:0061003 | positive regulation of dendritic spine morphogenesis | - | ISS | Process |
| GO:0061351 | neural precursor cell proliferation | - | IEA | Process |
| GO:0098978 | glutamatergic synapse | - | IEA | Component |
| GO:0099524 | postsynaptic cytosol | - | IEA | Component |
| GO:1902685 | positive regulation of receptor localization to synapse | - | ISS | Process |