
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 24746703 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| HNSC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THYM | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| KIRP | |||
| PAAD | |||
| TGCT | |||
| UCEC |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| MESO | ||
| HNSC | ||
| SKCM | ||
| ESCA | ||
| PAAD | ||
| BLCA | ||
| READ | ||
| LGG | ||
| LUAD |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000440086 | MMR_HSR1 | PF01926.23 | 2e-05 | 1 | 1 |
| ENSP00000264711 | MMR_HSR1 | PF01926.23 | 3.7e-05 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000534855 | DNAJC27-207 | 4831 | XM_011532902 | ENSP00000440086 | 177 (aa) | XP_011531204 | Q9NZQ0 |
| ENST00000380809 | DNAJC27-202 | 1378 | - | ENSP00000370187 | 90 (aa) | - | Q9NZQ0 |
| ENST00000468750 | DNAJC27-204 | 463 | - | - | - (aa) | - | - |
| ENST00000492985 | DNAJC27-205 | 485 | - | - | - (aa) | - | - |
| ENST00000494239 | DNAJC27-206 | 547 | - | - | - (aa) | - | - |
| ENST00000468467 | DNAJC27-203 | 561 | - | - | - (aa) | - | - |
| ENST00000264711 | DNAJC27-201 | 4992 | - | ENSP00000264711 | 273 (aa) | - | Q9NZQ0 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000115137 | rs4665736 | 2 | 24964730 | T | Height | 20881960 | [NR] unit increase | 0.029 | EFO_0004339 |
| ENSG00000115137 | rs1172294 | 2 | 24946331 | G | Pubertal anthropometrics | 23449627 | [0.053-0.107] unit decrease | 0.08 | EFO_0001382|EFO_0005201 |
| ENSG00000115137 | rs4665736 | 2 | 24964730 | T | Height | 25429064 | [0.023-0.051] unit increase | 0.037 | EFO_0004339 |
| ENSG00000115137 | rs1470039 | 2 | 24970600 | ? | Reaction time | 29844566 | [0.0046-0.0115] unit increase | 0.0080758 | EFO_0008393 |
| ENSG00000115137 | rs754537 | 2 | 24953408 | T | Body fat distribution (arm fat ratio) | 30664634 | NR unit decrease (replication) | 0.04375 | EFO_0004341 |
| ENSG00000115137 | rs754537 | 2 | 24953408 | T | Body fat distribution (arm fat ratio) | 30664634 | NR unit decrease (replication) | 0.02381 | EFO_0004341 |
| ENSG00000115137 | rs754537 | 2 | 24953408 | T | Body fat distribution (arm fat ratio) | 30664634 | NR unit increase (replication) | 4.25E-04 | EFO_0004341 |
| ENSG00000115137 | rs754537 | 2 | 24953408 | T | Body fat distribution (trunk fat ratio) | 30664634 | NR unit increase (replication) | 0.02357 | EFO_0004341 |
| ENSG00000115137 | rs754537 | 2 | 24953408 | T | Body fat distribution (trunk fat ratio) | 30664634 | NR unit increase (replication) | 0.01294 | EFO_0004341 |
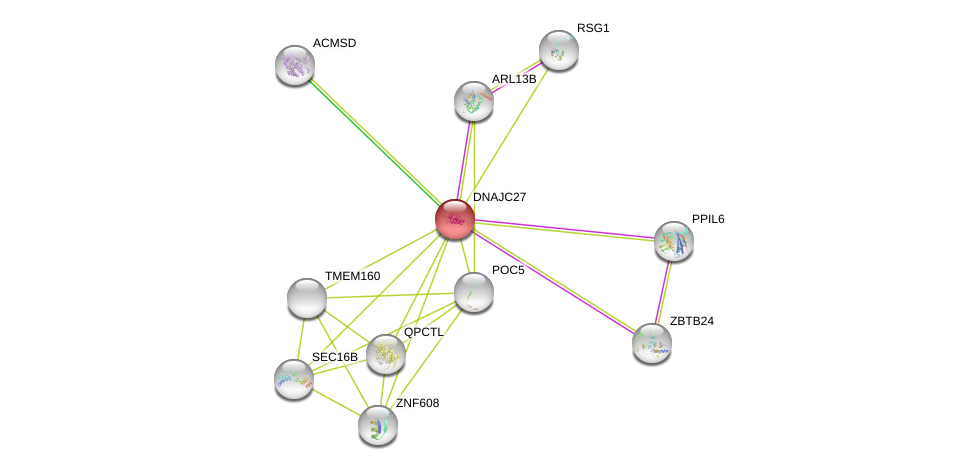
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000115137 | DNAJC27 | 100 | 82.784 | ENSAPOG00000020278 | dnajc27 | 100 | 82.784 | Acanthochromis_polyacanthus |
| ENSG00000115137 | DNAJC27 | 100 | 96.350 | ENSAMEG00000002554 | DNAJC27 | 100 | 96.350 | Ailuropoda_melanoleuca |
| ENSG00000115137 | DNAJC27 | 99 | 80.952 | ENSACIG00000009139 | dnajc27 | 100 | 80.952 | Amphilophus_citrinellus |
| ENSG00000115137 | DNAJC27 | 100 | 82.784 | ENSAOCG00000006604 | dnajc27 | 100 | 82.784 | Amphiprion_ocellaris |
| ENSG00000115137 | DNAJC27 | 100 | 82.784 | ENSAPEG00000009586 | dnajc27 | 100 | 82.784 | Amphiprion_percula |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSATEG00000008796 | dnajc27 | 100 | 82.418 | Anabas_testudineus |
| ENSG00000115137 | DNAJC27 | 100 | 89.011 | ENSACAG00000010248 | DNAJC27 | 100 | 89.011 | Anolis_carolinensis |
| ENSG00000115137 | DNAJC27 | 100 | 99.435 | ENSANAG00000032274 | DNAJC27 | 100 | 99.435 | Aotus_nancymaae |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSACLG00000004019 | dnajc27 | 100 | 82.418 | Astatotilapia_calliptera |
| ENSG00000115137 | DNAJC27 | 100 | 82.784 | ENSAMXG00000041488 | dnajc27 | 99 | 82.784 | Astyanax_mexicanus |
| ENSG00000115137 | DNAJC27 | 100 | 95.604 | ENSBTAG00000018239 | DNAJC27 | 100 | 95.604 | Bos_taurus |
| ENSG00000115137 | DNAJC27 | 100 | 99.435 | ENSCJAG00000008094 | DNAJC27 | 100 | 99.435 | Callithrix_jacchus |
| ENSG00000115137 | DNAJC27 | 100 | 96.350 | ENSCAFG00000004098 | DNAJC27 | 100 | 96.350 | Canis_familiaris |
| ENSG00000115137 | DNAJC27 | 100 | 97.070 | ENSCAFG00020025277 | DNAJC27 | 100 | 97.070 | Canis_lupus_dingo |
| ENSG00000115137 | DNAJC27 | 100 | 95.238 | ENSCHIG00000013821 | DNAJC27 | 100 | 95.238 | Capra_hircus |
| ENSG00000115137 | DNAJC27 | 100 | 98.305 | ENSTSYG00000014096 | DNAJC27 | 100 | 98.305 | Carlito_syrichta |
| ENSG00000115137 | DNAJC27 | 100 | 93.785 | ENSCPOG00000006843 | DNAJC27 | 100 | 93.407 | Cavia_porcellus |
| ENSG00000115137 | DNAJC27 | 100 | 98.535 | ENSCCAG00000021410 | DNAJC27 | 100 | 98.535 | Cebus_capucinus |
| ENSG00000115137 | DNAJC27 | 100 | 99.435 | ENSCATG00000029231 | DNAJC27 | 100 | 99.435 | Cercocebus_atys |
| ENSG00000115137 | DNAJC27 | 100 | 93.220 | ENSCLAG00000008212 | DNAJC27 | 100 | 92.674 | Chinchilla_lanigera |
| ENSG00000115137 | DNAJC27 | 100 | 99.634 | ENSCSAG00000008536 | DNAJC27 | 100 | 99.634 | Chlorocebus_sabaeus |
| ENSG00000115137 | DNAJC27 | 100 | 91.209 | ENSCPBG00000022284 | DNAJC27 | 100 | 91.209 | Chrysemys_picta_bellii |
| ENSG00000115137 | DNAJC27 | 98 | 67.052 | ENSCING00000000917 | rbj | 99 | 64.419 | Ciona_intestinalis |
| ENSG00000115137 | DNAJC27 | 96 | 70.930 | ENSCSAVG00000006409 | - | 99 | 65.543 | Ciona_savignyi |
| ENSG00000115137 | DNAJC27 | 100 | 99.435 | ENSCANG00000029144 | - | 100 | 99.435 | Colobus_angolensis_palliatus |
| ENSG00000115137 | DNAJC27 | 100 | 93.773 | ENSCGRG00001023794 | Dnajc27 | 100 | 93.773 | Cricetulus_griseus_chok1gshd |
| ENSG00000115137 | DNAJC27 | 100 | 93.773 | ENSCGRG00000007127 | Dnajc27 | 100 | 93.773 | Cricetulus_griseus_crigri |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSCSEG00000014284 | dnajc27 | 100 | 82.418 | Cynoglossus_semilaevis |
| ENSG00000115137 | DNAJC27 | 100 | 84.249 | ENSDARG00000070916 | dnajc27 | 100 | 84.249 | Danio_rerio |
| ENSG00000115137 | DNAJC27 | 100 | 96.350 | ENSDNOG00000040043 | DNAJC27 | 100 | 96.350 | Dasypus_novemcinctus |
| ENSG00000115137 | DNAJC27 | 100 | 96.703 | ENSDORG00000015134 | Dnajc27 | 100 | 96.703 | Dipodomys_ordii |
| ENSG00000115137 | DNAJC27 | 99 | 97.727 | ENSEASG00005010598 | DNAJC27 | 100 | 96.703 | Equus_asinus_asinus |
| ENSG00000115137 | DNAJC27 | 100 | 97.070 | ENSECAG00000024595 | DNAJC27 | 100 | 97.070 | Equus_caballus |
| ENSG00000115137 | DNAJC27 | 100 | 82.051 | ENSELUG00000019264 | dnajc27 | 100 | 82.051 | Esox_lucius |
| ENSG00000115137 | DNAJC27 | 100 | 97.175 | ENSFCAG00000038919 | DNAJC27 | 100 | 97.175 | Felis_catus |
| ENSG00000115137 | DNAJC27 | 91 | 89.024 | ENSFALG00000012658 | DNAJC27 | 100 | 75.636 | Ficedula_albicollis |
| ENSG00000115137 | DNAJC27 | 99 | 93.750 | ENSGALG00000028967 | DNAJC27 | 100 | 91.575 | Gallus_gallus |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSGACG00000015457 | dnajc27 | 100 | 82.418 | Gasterosteus_aculeatus |
| ENSG00000115137 | DNAJC27 | 100 | 99.634 | ENSGGOG00000004081 | DNAJC27 | 100 | 99.634 | Gorilla_gorilla |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSHBUG00000001491 | dnajc27 | 100 | 82.418 | Haplochromis_burtoni |
| ENSG00000115137 | DNAJC27 | 100 | 95.604 | ENSHGLG00000013756 | DNAJC27 | 100 | 95.604 | Heterocephalus_glaber_female |
| ENSG00000115137 | DNAJC27 | 100 | 95.604 | ENSHGLG00100009434 | DNAJC27 | 100 | 95.604 | Heterocephalus_glaber_male |
| ENSG00000115137 | DNAJC27 | 100 | 81.685 | ENSHCOG00000002948 | dnajc27 | 100 | 81.685 | Hippocampus_comes |
| ENSG00000115137 | DNAJC27 | 100 | 83.516 | ENSIPUG00000015086 | dnajc27 | 100 | 83.516 | Ictalurus_punctatus |
| ENSG00000115137 | DNAJC27 | 100 | 96.703 | ENSSTOG00000006080 | DNAJC27 | 100 | 96.703 | Ictidomys_tridecemlineatus |
| ENSG00000115137 | DNAJC27 | 100 | 94.139 | ENSJJAG00000016683 | Dnajc27 | 100 | 94.139 | Jaculus_jaculus |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSKMAG00000011954 | dnajc27 | 100 | 82.418 | Kryptolebias_marmoratus |
| ENSG00000115137 | DNAJC27 | 91 | 85.366 | ENSLBEG00000018738 | - | 95 | 66.543 | Labrus_bergylta |
| ENSG00000115137 | DNAJC27 | 100 | 82.051 | ENSLBEG00000028391 | dnajc27 | 100 | 82.051 | Labrus_bergylta |
| ENSG00000115137 | DNAJC27 | 100 | 82.784 | ENSLACG00000012994 | DNAJC27 | 100 | 82.784 | Latimeria_chalumnae |
| ENSG00000115137 | DNAJC27 | 100 | 83.051 | ENSLOCG00000016679 | dnajc27 | 100 | 82.784 | Lepisosteus_oculatus |
| ENSG00000115137 | DNAJC27 | 100 | 96.337 | ENSLAFG00000002742 | DNAJC27 | 100 | 96.337 | Loxodonta_africana |
| ENSG00000115137 | DNAJC27 | 100 | 99.435 | ENSMFAG00000043229 | DNAJC27 | 100 | 99.435 | Macaca_fascicularis |
| ENSG00000115137 | DNAJC27 | 100 | 99.435 | ENSMMUG00000009603 | DNAJC27 | 100 | 99.435 | Macaca_mulatta |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSMAMG00000022707 | dnajc27 | 100 | 82.418 | Mastacembelus_armatus |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSMZEG00005012058 | dnajc27 | 100 | 82.418 | Maylandia_zebra |
| ENSG00000115137 | DNAJC27 | 100 | 94.139 | ENSMAUG00000000099 | Dnajc27 | 100 | 94.139 | Mesocricetus_auratus |
| ENSG00000115137 | DNAJC27 | 100 | 98.870 | ENSMICG00000027424 | DNAJC27 | 100 | 98.870 | Microcebus_murinus |
| ENSG00000115137 | DNAJC27 | 100 | 93.796 | ENSMOCG00000015357 | Dnajc27 | 100 | 93.796 | Microtus_ochrogaster |
| ENSG00000115137 | DNAJC27 | 99 | 92.614 | ENSMODG00000015869 | DNAJC27 | 100 | 90.842 | Monodelphis_domestica |
| ENSG00000115137 | DNAJC27 | 100 | 82.051 | ENSMALG00000001189 | dnajc27 | 100 | 82.051 | Monopterus_albus |
| ENSG00000115137 | DNAJC27 | 100 | 93.407 | MGP_CAROLIEiJ_G0017579 | Dnajc27 | 100 | 93.407 | Mus_caroli |
| ENSG00000115137 | DNAJC27 | 100 | 93.407 | ENSMUSG00000020657 | Dnajc27 | 100 | 93.407 | Mus_musculus |
| ENSG00000115137 | DNAJC27 | 100 | 92.655 | MGP_PahariEiJ_G0029339 | - | 81 | 92.655 | Mus_pahari |
| ENSG00000115137 | DNAJC27 | 100 | 93.407 | MGP_SPRETEiJ_G0018423 | Dnajc27 | 100 | 93.407 | Mus_spretus |
| ENSG00000115137 | DNAJC27 | 100 | 96.703 | ENSMPUG00000014032 | DNAJC27 | 100 | 96.703 | Mustela_putorius_furo |
| ENSG00000115137 | DNAJC27 | 100 | 93.773 | ENSMLUG00000014143 | DNAJC27 | 100 | 93.773 | Myotis_lucifugus |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSNBRG00000022552 | dnajc27 | 100 | 82.418 | Neolamprologus_brichardi |
| ENSG00000115137 | DNAJC27 | 100 | 100.000 | ENSNLEG00000001103 | DNAJC27 | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000115137 | DNAJC27 | 99 | 94.886 | ENSOPRG00000008650 | DNAJC27 | 100 | 93.407 | Ochotona_princeps |
| ENSG00000115137 | DNAJC27 | 100 | 93.220 | ENSODEG00000007488 | DNAJC27 | 100 | 93.040 | Octodon_degus |
| ENSG00000115137 | DNAJC27 | 100 | 80.791 | ENSONIG00000009008 | dnajc27 | 97 | 82.061 | Oreochromis_niloticus |
| ENSG00000115137 | DNAJC27 | 100 | 96.703 | ENSOCUG00000011260 | DNAJC27 | 100 | 96.703 | Oryctolagus_cuniculus |
| ENSG00000115137 | DNAJC27 | 100 | 81.319 | ENSORLG00000001134 | dnajc27 | 100 | 81.319 | Oryzias_latipes |
| ENSG00000115137 | DNAJC27 | 100 | 81.319 | ENSORLG00020006790 | dnajc27 | 100 | 81.319 | Oryzias_latipes_hni |
| ENSG00000115137 | DNAJC27 | 100 | 81.319 | ENSORLG00015010005 | dnajc27 | 100 | 81.319 | Oryzias_latipes_hsok |
| ENSG00000115137 | DNAJC27 | 100 | 81.319 | ENSOMEG00000017890 | dnajc27 | 100 | 81.319 | Oryzias_melastigma |
| ENSG00000115137 | DNAJC27 | 100 | 96.337 | ENSOGAG00000015168 | DNAJC27 | 100 | 96.337 | Otolemur_garnettii |
| ENSG00000115137 | DNAJC27 | 100 | 94.891 | ENSOARG00000018127 | DNAJC27 | 100 | 94.891 | Ovis_aries |
| ENSG00000115137 | DNAJC27 | 100 | 100.000 | ENSPPAG00000031678 | DNAJC27 | 100 | 100.000 | Pan_paniscus |
| ENSG00000115137 | DNAJC27 | 100 | 96.703 | ENSPPRG00000022920 | DNAJC27 | 100 | 96.703 | Panthera_pardus |
| ENSG00000115137 | DNAJC27 | 100 | 96.703 | ENSPTIG00000013443 | DNAJC27 | 100 | 96.703 | Panthera_tigris_altaica |
| ENSG00000115137 | DNAJC27 | 100 | 100.000 | ENSPTRG00000011719 | DNAJC27 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000115137 | DNAJC27 | 100 | 98.901 | ENSPANG00000023972 | DNAJC27 | 100 | 98.901 | Papio_anubis |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSPKIG00000005931 | DNAJC27 | 100 | 82.418 | Paramormyrops_kingsleyae |
| ENSG00000115137 | DNAJC27 | 99 | 82.386 | ENSPKIG00000005039 | dnajc27 | 96 | 82.386 | Paramormyrops_kingsleyae |
| ENSG00000115137 | DNAJC27 | 99 | 92.614 | ENSPSIG00000009930 | DNAJC27 | 100 | 91.209 | Pelodiscus_sinensis |
| ENSG00000115137 | DNAJC27 | 98 | 78.035 | ENSPMAG00000005573 | dnajc27 | 98 | 75.458 | Petromyzon_marinus |
| ENSG00000115137 | DNAJC27 | 99 | 93.182 | ENSPCIG00000025239 | DNAJC27 | 100 | 91.941 | Phascolarctos_cinereus |
| ENSG00000115137 | DNAJC27 | 100 | 81.685 | ENSPFOG00000006335 | dnajc27 | 100 | 81.685 | Poecilia_formosa |
| ENSG00000115137 | DNAJC27 | 100 | 82.051 | ENSPLAG00000008845 | dnajc27 | 100 | 82.051 | Poecilia_latipinna |
| ENSG00000115137 | DNAJC27 | 100 | 81.685 | ENSPMEG00000018256 | dnajc27 | 100 | 81.685 | Poecilia_mexicana |
| ENSG00000115137 | DNAJC27 | 99 | 80.114 | ENSPREG00000010228 | dnajc27 | 93 | 78.453 | Poecilia_reticulata |
| ENSG00000115137 | DNAJC27 | 100 | 100.000 | ENSPPYG00000012594 | DNAJC27 | 100 | 100.000 | Pongo_abelii |
| ENSG00000115137 | DNAJC27 | 100 | 98.305 | ENSPCOG00000025665 | DNAJC27 | 100 | 98.305 | Propithecus_coquereli |
| ENSG00000115137 | DNAJC27 | 100 | 97.802 | ENSPVAG00000017520 | DNAJC27 | 100 | 97.802 | Pteropus_vampyrus |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSPNYG00000023681 | dnajc27 | 100 | 82.418 | Pundamilia_nyererei |
| ENSG00000115137 | DNAJC27 | 100 | 82.784 | ENSPNAG00000025619 | dnajc27 | 100 | 82.784 | Pygocentrus_nattereri |
| ENSG00000115137 | DNAJC27 | 100 | 93.040 | ENSRNOG00000003988 | Dnajc27 | 100 | 93.040 | Rattus_norvegicus |
| ENSG00000115137 | DNAJC27 | 100 | 98.870 | ENSRBIG00000041935 | - | 100 | 98.870 | Rhinopithecus_bieti |
| ENSG00000115137 | DNAJC27 | 100 | 99.435 | ENSRROG00000031027 | - | 100 | 99.435 | Rhinopithecus_roxellana |
| ENSG00000115137 | DNAJC27 | 91 | 93.902 | ENSSHAG00000002151 | - | 93 | 92.647 | Sarcophilus_harrisii |
| ENSG00000115137 | DNAJC27 | 100 | 83.150 | ENSSFOG00015017512 | dnajc27 | 100 | 83.150 | Scleropages_formosus |
| ENSG00000115137 | DNAJC27 | 100 | 83.150 | ENSSFOG00015020074 | DNAJC27 | 100 | 83.150 | Scleropages_formosus |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSSMAG00000000032 | dnajc27 | 100 | 82.418 | Scophthalmus_maximus |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSSDUG00000003036 | dnajc27 | 100 | 82.418 | Seriola_dumerili |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSSLDG00000015143 | dnajc27 | 100 | 82.418 | Seriola_lalandi_dorsalis |
| ENSG00000115137 | DNAJC27 | 91 | 90.244 | ENSSPUG00000006583 | DNAJC27 | 99 | 74.214 | Sphenodon_punctatus |
| ENSG00000115137 | DNAJC27 | 100 | 82.418 | ENSSPAG00000009154 | dnajc27 | 100 | 82.418 | Stegastes_partitus |
| ENSG00000115137 | DNAJC27 | 100 | 96.610 | ENSSSCG00000008577 | DNAJC27 | 100 | 96.610 | Sus_scrofa |
| ENSG00000115137 | DNAJC27 | 100 | 82.784 | ENSTRUG00000023427 | dnajc27 | 100 | 82.784 | Takifugu_rubripes |
| ENSG00000115137 | DNAJC27 | 100 | 82.784 | ENSTNIG00000006596 | dnajc27 | 100 | 82.784 | Tetraodon_nigroviridis |
| ENSG00000115137 | DNAJC27 | 100 | 97.436 | ENSTBEG00000005463 | DNAJC27 | 100 | 97.436 | Tupaia_belangeri |
| ENSG00000115137 | DNAJC27 | 100 | 95.604 | ENSTTRG00000015163 | DNAJC27 | 100 | 95.604 | Tursiops_truncatus |
| ENSG00000115137 | DNAJC27 | 100 | 97.070 | ENSUAMG00000009585 | DNAJC27 | 100 | 97.070 | Ursus_americanus |
| ENSG00000115137 | DNAJC27 | 100 | 97.070 | ENSUMAG00000022558 | DNAJC27 | 100 | 97.070 | Ursus_maritimus |
| ENSG00000115137 | DNAJC27 | 100 | 89.011 | ENSVPAG00000006953 | DNAJC27 | 100 | 89.011 | Vicugna_pacos |
| ENSG00000115137 | DNAJC27 | 100 | 96.703 | ENSVVUG00000015241 | DNAJC27 | 100 | 96.703 | Vulpes_vulpes |
| ENSG00000115137 | DNAJC27 | 100 | 81.319 | ENSXCOG00000010812 | dnajc27 | 100 | 81.319 | Xiphophorus_couchianus |
| ENSG00000115137 | DNAJC27 | 100 | 81.319 | ENSXMAG00000003407 | dnajc27 | 100 | 81.319 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003924 | GTPase activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 25416956. | IPI | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005634 | nucleus | - | IEA | Component |
| GO:0006886 | intracellular protein transport | 21873635. | IBA | Process |
| GO:0032482 | Rab protein signal transduction | 21873635. | IBA | Process |
| GO:0070374 | positive regulation of ERK1 and ERK2 cascade | - | IEA | Process |
| GO:0071701 | regulation of MAPK export from nucleus | - | IEA | Process |