
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| GBM | |||
| KIRC | |||
| LGG | |||
| MESO | |||
| PAAD | |||
| SKCM | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| ACC | ||
| SKCM | ||
| BLCA | ||
| READ | ||
| LAML | ||
| GBM | ||
| LGG |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000441048 | AKAP7_NLS | PF10469.9 | 4.4e-47 | 1 | 1 |
| ENSP00000405252 | AKAP7_NLS | PF10469.9 | 4.6e-47 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000474850 | AKAP7-205 | 670 | - | ENSP00000418208 | 104 (aa) | - | O43687 |
| ENST00000537868 | AKAP7-207 | 1006 | - | ENSP00000446022 | 87 (aa) | - | F5H4P9 |
| ENST00000342266 | AKAP7-202 | 2461 | - | ENSP00000345149 | 81 (aa) | - | O43687 |
| ENST00000431975 | AKAP7-204 | 2954 | XM_006715614 | ENSP00000405252 | 348 (aa) | XP_006715677 | Q9P0M2 |
| ENST00000543815 | AKAP7-209 | 412 | - | - | - (aa) | - | - |
| ENST00000263050 | AKAP7-201 | 2238 | - | ENSP00000263050 | 84 (aa) | - | J3KN37 |
| ENST00000366358 | AKAP7-203 | 901 | - | - | - (aa) | - | - |
| ENST00000535150 | AKAP7-206 | 1041 | - | - | - (aa) | - | - |
| ENST00000541650 | AKAP7-208 | 2008 | XM_005267228 | ENSP00000441048 | 350 (aa) | XP_005267285 | F5GXD1 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000118507 | Carotid Stenosis | 1.9286045692235E-5 | 17903303 |
| ENSG00000118507 | Osteoporosis | 4.70131377372507E-5 | 17903296 |
| ENSG00000118507 | Heart Failure | 8.2837975E-004 | - |
| ENSG00000118507 | Alcoholism | 7.5236e-005 | 22978509 |
| ENSG00000118507 | Bilirubin | 1.024e-005 | - |
| ENSG00000118507 | Bilirubin | 1.024e-005 | - |
| ENSG00000118507 | Arteries | 3.0600000E-005 | - |
| ENSG00000118507 | Child Development Disorders, Pervasive | 3.4000000E-005 | - |
| ENSG00000118507 | C-Reactive Protein | 3E-8 | 24763700 |
| ENSG00000118507 | Multiple Myeloma | 6E-6 | 23502783 |
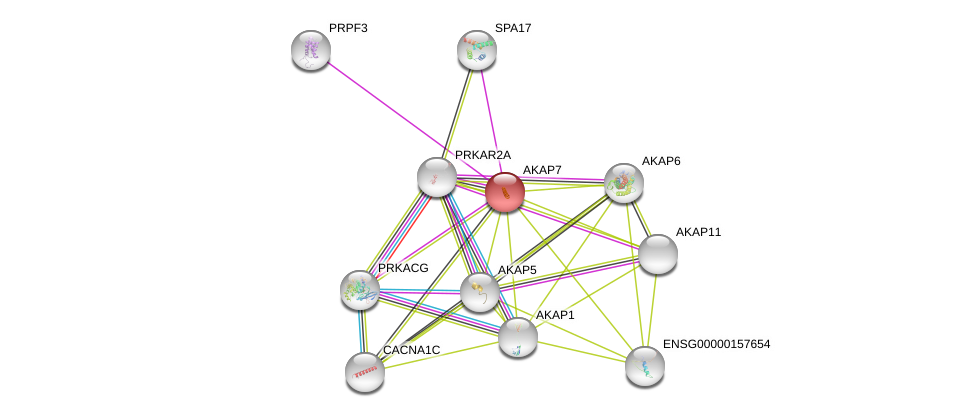
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000118507 | AKAP7 | 80 | 83.871 | ENSAMEG00000007663 | AKAP7 | 100 | 83.871 | Ailuropoda_melanoleuca |
| ENSG00000118507 | AKAP7 | 74 | 56.322 | ENSAPLG00000010992 | AKAP7 | 99 | 56.705 | Anas_platyrhynchos |
| ENSG00000118507 | AKAP7 | 71 | 53.571 | ENSACAG00000001820 | AKAP7 | 92 | 55.645 | Anolis_carolinensis |
| ENSG00000118507 | AKAP7 | 100 | 96.429 | ENSANAG00000020367 | AKAP7 | 100 | 90.805 | Aotus_nancymaae |
| ENSG00000118507 | AKAP7 | 85 | 56.338 | ENSAMXG00000010500 | AKAP7 | 84 | 39.683 | Astyanax_mexicanus |
| ENSG00000118507 | AKAP7 | 100 | 85.057 | ENSBTAG00000000240 | AKAP7 | 100 | 82.371 | Bos_taurus |
| ENSG00000118507 | AKAP7 | 100 | 97.619 | ENSCJAG00000003157 | AKAP7 | 100 | 92.529 | Callithrix_jacchus |
| ENSG00000118507 | AKAP7 | 98 | 79.268 | ENSCAFG00000001147 | AKAP7 | 96 | 79.938 | Canis_familiaris |
| ENSG00000118507 | AKAP7 | 100 | 86.207 | ENSCHIG00000020750 | AKAP7 | 100 | 82.675 | Capra_hircus |
| ENSG00000118507 | AKAP7 | 100 | 88.095 | ENSTSYG00000028287 | AKAP7 | 99 | 87.941 | Carlito_syrichta |
| ENSG00000118507 | AKAP7 | 100 | 82.143 | ENSCPOG00000035986 | AKAP7 | 99 | 73.731 | Cavia_porcellus |
| ENSG00000118507 | AKAP7 | 100 | 89.655 | ENSCCAG00000026328 | - | 100 | 85.470 | Cebus_capucinus |
| ENSG00000118507 | AKAP7 | 100 | 97.619 | ENSCCAG00000035834 | AKAP7 | 100 | 92.816 | Cebus_capucinus |
| ENSG00000118507 | AKAP7 | 100 | 98.810 | ENSCATG00000029710 | AKAP7 | 100 | 96.552 | Cercocebus_atys |
| ENSG00000118507 | AKAP7 | 100 | 83.908 | ENSCLAG00000008875 | AKAP7 | 99 | 76.627 | Chinchilla_lanigera |
| ENSG00000118507 | AKAP7 | 100 | 97.619 | ENSCSAG00000016634 | AKAP7 | 100 | 96.552 | Chlorocebus_sabaeus |
| ENSG00000118507 | AKAP7 | 75 | 83.142 | ENSCHOG00000009698 | AKAP7 | 80 | 83.142 | Choloepus_hoffmanni |
| ENSG00000118507 | AKAP7 | 95 | 79.221 | ENSCPBG00000018259 | AKAP7 | 76 | 68.582 | Chrysemys_picta_bellii |
| ENSG00000118507 | AKAP7 | 60 | 90.476 | ENSCANG00000040046 | AKAP7 | 97 | 90.476 | Colobus_angolensis_palliatus |
| ENSG00000118507 | AKAP7 | 100 | 82.143 | ENSCGRG00001014402 | Akap7 | 100 | 81.672 | Cricetulus_griseus_chok1gshd |
| ENSG00000118507 | AKAP7 | 100 | 82.143 | ENSCGRG00000000308 | Akap7 | 100 | 81.672 | Cricetulus_griseus_crigri |
| ENSG00000118507 | AKAP7 | 61 | 81.991 | ENSDNOG00000003557 | AKAP7 | 97 | 81.991 | Dasypus_novemcinctus |
| ENSG00000118507 | AKAP7 | 96 | 80.247 | ENSETEG00000013023 | AKAP7 | 98 | 75.087 | Echinops_telfairi |
| ENSG00000118507 | AKAP7 | 80 | 77.419 | ENSEASG00005009720 | AKAP7 | 90 | 77.419 | Equus_asinus_asinus |
| ENSG00000118507 | AKAP7 | 100 | 82.759 | ENSECAG00000022011 | AKAP7 | 100 | 80.547 | Equus_caballus |
| ENSG00000118507 | AKAP7 | 100 | 84.524 | ENSEEUG00000010380 | AKAP7 | 63 | 81.463 | Erinaceus_europaeus |
| ENSG00000118507 | AKAP7 | 98 | 54.878 | ENSELUG00000020202 | AKAP7 | 75 | 45.245 | Esox_lucius |
| ENSG00000118507 | AKAP7 | 100 | 89.286 | ENSFCAG00000012005 | AKAP7 | 100 | 82.857 | Felis_catus |
| ENSG00000118507 | AKAP7 | 72 | 56.972 | ENSFALG00000011697 | AKAP7 | 100 | 56.972 | Ficedula_albicollis |
| ENSG00000118507 | AKAP7 | 98 | 70.732 | ENSGALG00000029794 | AKAP7 | 68 | 58.255 | Gallus_gallus |
| ENSG00000118507 | AKAP7 | 95 | 81.250 | ENSGAGG00000012287 | AKAP7 | 94 | 52.836 | Gopherus_agassizii |
| ENSG00000118507 | AKAP7 | 100 | 99.713 | ENSGGOG00000008778 | AKAP7 | 100 | 99.713 | Gorilla_gorilla |
| ENSG00000118507 | AKAP7 | 99 | 77.907 | ENSHGLG00000015638 | AKAP7 | 90 | 77.019 | Heterocephalus_glaber_female |
| ENSG00000118507 | AKAP7 | 99 | 77.907 | ENSHGLG00100014062 | AKAP7 | 97 | 77.019 | Heterocephalus_glaber_male |
| ENSG00000118507 | AKAP7 | 92 | 37.500 | ENSIPUG00000014244 | akap7 | 76 | 37.764 | Ictalurus_punctatus |
| ENSG00000118507 | AKAP7 | 100 | 85.057 | ENSSTOG00000016147 | AKAP7 | 99 | 83.871 | Ictidomys_tridecemlineatus |
| ENSG00000118507 | AKAP7 | 100 | 82.143 | ENSJJAG00000021718 | Akap7 | 100 | 83.108 | Jaculus_jaculus |
| ENSG00000118507 | AKAP7 | 94 | 70.886 | ENSLACG00000011179 | AKAP7 | 77 | 50.150 | Latimeria_chalumnae |
| ENSG00000118507 | AKAP7 | 73 | 54.086 | ENSLOCG00000017275 | AKAP7 | 80 | 53.184 | Lepisosteus_oculatus |
| ENSG00000118507 | AKAP7 | 70 | 79.012 | ENSLAFG00000005191 | AKAP7 | 98 | 78.601 | Loxodonta_africana |
| ENSG00000118507 | AKAP7 | 100 | 98.810 | ENSMFAG00000044065 | AKAP7 | 100 | 96.839 | Macaca_fascicularis |
| ENSG00000118507 | AKAP7 | 100 | 98.810 | ENSMMUG00000000577 | AKAP7 | 100 | 96.839 | Macaca_mulatta |
| ENSG00000118507 | AKAP7 | 100 | 98.810 | ENSMNEG00000031192 | AKAP7 | 100 | 96.839 | Macaca_nemestrina |
| ENSG00000118507 | AKAP7 | 100 | 97.619 | ENSMLEG00000001626 | AKAP7 | 100 | 95.977 | Mandrillus_leucophaeus |
| ENSG00000118507 | AKAP7 | 74 | 55.385 | ENSMGAG00000013177 | AKAP7 | 99 | 55.556 | Meleagris_gallopavo |
| ENSG00000118507 | AKAP7 | 100 | 80.952 | ENSMAUG00000012136 | Akap7 | 100 | 81.350 | Mesocricetus_auratus |
| ENSG00000118507 | AKAP7 | 100 | 91.667 | ENSMICG00000015262 | AKAP7 | 100 | 85.714 | Microcebus_murinus |
| ENSG00000118507 | AKAP7 | 100 | 79.762 | ENSMOCG00000012192 | Akap7 | 100 | 80.707 | Microtus_ochrogaster |
| ENSG00000118507 | AKAP7 | 100 | 78.824 | ENSMODG00000017586 | - | 66 | 68.678 | Monodelphis_domestica |
| ENSG00000118507 | AKAP7 | 100 | 80.952 | MGP_CAROLIEiJ_G0015193 | Akap7 | 100 | 78.457 | Mus_caroli |
| ENSG00000118507 | AKAP7 | 100 | 80.952 | ENSMUSG00000039166 | Akap7 | 100 | 78.981 | Mus_musculus |
| ENSG00000118507 | AKAP7 | 100 | 79.762 | MGP_PahariEiJ_G0023815 | Akap7 | 100 | 78.778 | Mus_pahari |
| ENSG00000118507 | AKAP7 | 100 | 80.952 | MGP_SPRETEiJ_G0016002 | Akap7 | 100 | 79.100 | Mus_spretus |
| ENSG00000118507 | AKAP7 | 100 | 86.905 | ENSMPUG00000014649 | AKAP7 | 100 | 84.543 | Mustela_putorius_furo |
| ENSG00000118507 | AKAP7 | 99 | 86.747 | ENSMLUG00000006443 | AKAP7 | 99 | 82.596 | Myotis_lucifugus |
| ENSG00000118507 | AKAP7 | 100 | 83.333 | ENSNGAG00000020889 | Akap7 | 100 | 82.958 | Nannospalax_galili |
| ENSG00000118507 | AKAP7 | 100 | 98.810 | ENSNLEG00000014330 | AKAP7 | 100 | 98.276 | Nomascus_leucogenys |
| ENSG00000118507 | AKAP7 | 100 | 75.294 | ENSMEUG00000001176 | - | 100 | 63.492 | Notamacropus_eugenii |
| ENSG00000118507 | AKAP7 | 67 | 81.466 | ENSOPRG00000015646 | AKAP7 | 100 | 81.466 | Ochotona_princeps |
| ENSG00000118507 | AKAP7 | 100 | 85.714 | ENSOCUG00000013991 | AKAP7 | 99 | 84.498 | Oryctolagus_cuniculus |
| ENSG00000118507 | AKAP7 | 100 | 82.759 | ENSOGAG00000010862 | AKAP7 | 100 | 84.722 | Otolemur_garnettii |
| ENSG00000118507 | AKAP7 | 79 | 81.884 | ENSOARG00000013910 | AKAP7 | 98 | 81.884 | Ovis_aries |
| ENSG00000118507 | AKAP7 | 100 | 100.000 | ENSPPAG00000039914 | AKAP7 | 100 | 99.425 | Pan_paniscus |
| ENSG00000118507 | AKAP7 | 100 | 98.851 | ENSPPAG00000006789 | - | 100 | 99.128 | Pan_paniscus |
| ENSG00000118507 | AKAP7 | 100 | 89.286 | ENSPPRG00000005682 | AKAP7 | 100 | 82.000 | Panthera_pardus |
| ENSG00000118507 | AKAP7 | 100 | 89.286 | ENSPTIG00000013299 | AKAP7 | 98 | 82.216 | Panthera_tigris_altaica |
| ENSG00000118507 | AKAP7 | 100 | 100.000 | ENSPTRG00000018599 | AKAP7 | 100 | 99.138 | Pan_troglodytes |
| ENSG00000118507 | AKAP7 | 100 | 98.810 | ENSPANG00000023385 | AKAP7 | 100 | 96.264 | Papio_anubis |
| ENSG00000118507 | AKAP7 | 86 | 58.228 | ENSPKIG00000012538 | AKAP7 | 83 | 48.065 | Paramormyrops_kingsleyae |
| ENSG00000118507 | AKAP7 | 98 | 78.313 | ENSPSIG00000011783 | AKAP7 | 99 | 60.883 | Pelodiscus_sinensis |
| ENSG00000118507 | AKAP7 | 100 | 79.762 | ENSPEMG00000021361 | Akap7 | 100 | 78.457 | Peromyscus_maniculatus_bairdii |
| ENSG00000118507 | AKAP7 | 84 | 42.517 | ENSPMAG00000006828 | AKAP7 | 100 | 38.438 | Petromyzon_marinus |
| ENSG00000118507 | AKAP7 | 100 | 80.000 | ENSPCIG00000020677 | - | 63 | 72.843 | Phascolarctos_cinereus |
| ENSG00000118507 | AKAP7 | 100 | 100.000 | ENSPPYG00000017006 | AKAP7 | 100 | 99.387 | Pongo_abelii |
| ENSG00000118507 | AKAP7 | 79 | 76.562 | ENSPCAG00000016790 | AKAP7 | 100 | 58.282 | Procavia_capensis |
| ENSG00000118507 | AKAP7 | 100 | 92.857 | ENSPCOG00000016830 | AKAP7 | 100 | 71.552 | Propithecus_coquereli |
| ENSG00000118507 | AKAP7 | 100 | 88.095 | ENSPVAG00000005454 | AKAP7 | 100 | 86.196 | Pteropus_vampyrus |
| ENSG00000118507 | AKAP7 | 85 | 57.746 | ENSPNAG00000015970 | AKAP7 | 78 | 43.402 | Pygocentrus_nattereri |
| ENSG00000118507 | AKAP7 | 100 | 82.143 | ENSRNOG00000013202 | Akap7 | 100 | 73.295 | Rattus_norvegicus |
| ENSG00000118507 | AKAP7 | 100 | 98.810 | ENSRBIG00000029419 | AKAP7 | 100 | 96.264 | Rhinopithecus_bieti |
| ENSG00000118507 | AKAP7 | 81 | 95.760 | ENSRROG00000007555 | AKAP7 | 100 | 95.760 | Rhinopithecus_roxellana |
| ENSG00000118507 | AKAP7 | 100 | 96.429 | ENSSBOG00000024938 | AKAP7 | 100 | 91.789 | Saimiri_boliviensis_boliviensis |
| ENSG00000118507 | AKAP7 | 73 | 72.047 | ENSSHAG00000001178 | AKAP7 | 100 | 72.047 | Sarcophilus_harrisii |
| ENSG00000118507 | AKAP7 | 70 | 67.797 | ENSSFOG00015017908 | AKAP7 | 72 | 48.860 | Scleropages_formosus |
| ENSG00000118507 | AKAP7 | 100 | 76.190 | ENSSARG00000002384 | - | 72 | 75.439 | Sorex_araneus |
| ENSG00000118507 | AKAP7 | 63 | 50.455 | ENSSPUG00000008320 | AKAP7 | 71 | 50.455 | Sphenodon_punctatus |
| ENSG00000118507 | AKAP7 | 100 | 91.667 | ENSSSCG00000036790 | AKAP7 | 81 | 82.184 | Sus_scrofa |
| ENSG00000118507 | AKAP7 | 98 | 72.289 | ENSTGUG00000011679 | AKAP7 | 96 | 64.151 | Taeniopygia_guttata |
| ENSG00000118507 | AKAP7 | 100 | 90.476 | ENSTBEG00000010053 | AKAP7 | 100 | 84.969 | Tupaia_belangeri |
| ENSG00000118507 | AKAP7 | 100 | 91.667 | ENSTTRG00000016304 | AKAP7 | 100 | 77.301 | Tursiops_truncatus |
| ENSG00000118507 | AKAP7 | 100 | 86.207 | ENSUMAG00000000235 | AKAP7 | 100 | 82.812 | Ursus_maritimus |
| ENSG00000118507 | AKAP7 | 75 | 74.615 | ENSVPAG00000007961 | AKAP7 | 99 | 74.615 | Vicugna_pacos |
| ENSG00000118507 | AKAP7 | 98 | 79.268 | ENSVVUG00000001861 | AKAP7 | 96 | 80.495 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | - | IEA | Function |
| GO:0005515 | protein binding | 17244820. | IPI | Function |
| GO:0005634 | nucleus | - | IEA | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | 10613906. | IDA | Component |
| GO:0008150 | biological_process | - | ND | Process |
| GO:0010738 | regulation of protein kinase A signaling | 21873635. | IBA | Process |
| GO:0019901 | protein kinase binding | - | IEA | Function |
| GO:0032991 | protein-containing complex | 25097019. | IDA | Component |
| GO:0034237 | protein kinase A regulatory subunit binding | 21873635. | IBA | Function |
| GO:0050804 | modulation of chemical synaptic transmission | - | IEA | Process |
| GO:0051018 | protein kinase A binding | 10613906. | IDA | Function |
| GO:0051018 | protein kinase A binding | 25097019. | IPI | Function |
| GO:0098686 | hippocampal mossy fiber to CA3 synapse | - | IEA | Component |