
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CHOL | |||
| COAD | |||
| MESO | |||
| OV | |||
| PAAD | |||
| READ | |||
| THCA | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| ACC | ||
| HNSC | ||
| SKCM | ||
| PRAD | ||
| KIRP | ||
| CESC | ||
| LAML | ||
| LGG | ||
| LUAD | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000370508 | GOT1-201 | 2923 | - | ENSP00000359539 | 413 (aa) | - | P17174 |
| ENST00000471741 | GOT1-202 | 716 | - | - | - (aa) | - | - |
| ENST00000489349 | GOT1-203 | 646 | - | - | - (aa) | - | - |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00220 | Arginine biosynthesis | KEGG |
| hsa00250 | Alanine, aspartate and glutamate metabolism | KEGG |
| hsa00270 | Cysteine and methionine metabolism | KEGG |
| hsa00330 | Arginine and proline metabolism | KEGG |
| hsa00350 | Tyrosine metabolism | KEGG |
| hsa00360 | Phenylalanine metabolism | KEGG |
| hsa00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | KEGG |
| hsa01100 | Metabolic pathways | KEGG |
| hsa01200 | Carbon metabolism | KEGG |
| hsa01210 | 2-Oxocarboxylic acid metabolism | KEGG |
| hsa01230 | Biosynthesis of amino acids | KEGG |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000120053 | Heart Failure | 7.7333047E-004 | - |
| ENSG00000120053 | Platelet Function Tests | 1.3614200E-005 | - |
| ENSG00000120053 | Dental Caries | 6E-6 | 23064961 |
| ENSG00000120053 | Inflammatory Bowel Diseases | 2E-34 | 28067908 |
| ENSG00000120053 | Crohn Disease | 2E-20 | 21102463 |
| ENSG00000120053 | Inflammatory Bowel Diseases | 1E-54 | 23128233 |
| ENSG00000120053 | Erythrocyte Indices | 1E-10 | 23222517 |
| ENSG00000120053 | Crohn Disease | 1E-36 | 26192919 |
| ENSG00000120053 | Hypothyroidism | 1E-10 | 27182965 |
| ENSG00000120053 | Inflammatory Bowel Diseases | 2E-63 | 26192919 |
| ENSG00000120053 | Crohn Disease | 6E-47 | 26192919 |
| ENSG00000120053 | Aspartate Aminotransferases | 9E-7 | 24124411 |
| ENSG00000120053 | Colitis, Ulcerative | 2E-36 | 26192919 |
| ENSG00000120053 | Crohn Disease | 2E-24 | 28067908 |
| ENSG00000120053 | Colitis, Ulcerative | 2E-21 | 28067908 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000120053 | rs76850691 | 10 | 99397681 | C | Liver enzyme levels (aspartate transaminase) | 24124411 | [0.06-0.138] unit decrease | 0.099 | EFO_0004736 |
| ENSG00000120053 | rs11819158 | 10 | 99418299 | ? | Peripheral arterial disease (traffic-related air pollution interaction) | 27082954 | [6.5-9.6] | 8.05 | EFO_0004265|EFO_0007908 |
| ENSG00000120053 | rs11813054 | 10 | 99420879 | ? | Adolescent idiopathic scoliosis | 30019117 | EFO_0005423 | ||
| ENSG00000120053 | rs76850691 | 10 | 99397681 | C | Aspartate aminotransferase levels | 30718733 | [0.1-0.2] unit decrease | 0.151 | EFO_0004736 |
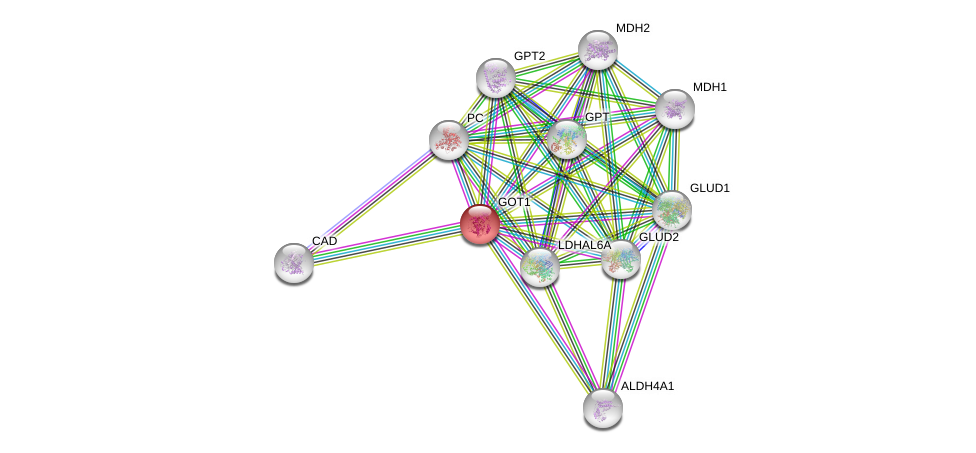
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000120053 | GOT1 | 99 | 53.790 | WBGene00020146 | got-1.2 | 99 | 53.790 | Caenorhabditis_elegans |
| ENSG00000120053 | GOT1 | 98 | 56.404 | FBgn0001124 | Got1 | 96 | 56.404 | Drosophila_melanogaster |
| ENSG00000120053 | GOT1 | 98 | 47.407 | FBgn0001125 | Got2 | 94 | 47.407 | Drosophila_melanogaster |
| ENSG00000120053 | GOT1 | 98 | 48.889 | ENSG00000125166 | GOT2 | 93 | 48.894 | Homo_sapiens |
| ENSG00000120053 | GOT1 | 98 | 48.642 | ENSMUSG00000031672 | Got2 | 93 | 48.649 | Mus_musculus |
| ENSG00000120053 | GOT1 | 98 | 46.602 | YLR027C | AAT2 | 97 | 46.602 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity | 21873635. | IBA | Function |
| GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity | 2182221.2241899.2731362.6391741. | IDA | Function |
| GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity | - | ISS | Function |
| GO:0004609 | phosphatidylserine decarboxylase activity | - | IEA | Function |
| GO:0005634 | nucleus | 21630459. | HDA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005737 | cytoplasm | 7060339. | IDA | Component |
| GO:0005764 | lysosome | - | IEA | Component |
| GO:0005829 | cytosol | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006094 | gluconeogenesis | - | TAS | Process |
| GO:0006103 | 2-oxoglutarate metabolic process | - | ISS | Process |
| GO:0006107 | oxaloacetate metabolic process | - | IEA | Process |
| GO:0006114 | glycerol biosynthetic process | - | ISS | Process |
| GO:0006531 | aspartate metabolic process | - | ISS | Process |
| GO:0006532 | aspartate biosynthetic process | 21873635. | IBA | Process |
| GO:0006533 | aspartate catabolic process | 2241899. | IDA | Process |
| GO:0006536 | glutamate metabolic process | - | ISS | Process |
| GO:0007219 | Notch signaling pathway | - | IEA | Process |
| GO:0008652 | cellular amino acid biosynthetic process | - | TAS | Process |
| GO:0009743 | response to carbohydrate | - | IEA | Process |
| GO:0019550 | glutamate catabolic process to aspartate | - | IEA | Process |
| GO:0019551 | glutamate catabolic process to 2-oxoglutarate | - | IEA | Process |
| GO:0030170 | pyridoxal phosphate binding | - | IEA | Function |
| GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | - | IEA | Process |
| GO:0031406 | carboxylic acid binding | - | IEA | Function |
| GO:0032869 | cellular response to insulin stimulus | 8396422. | IEP | Process |
| GO:0032966 | negative regulation of collagen biosynthetic process | - | IEA | Process |
| GO:0035902 | response to immobilization stress | - | IEA | Process |
| GO:0043679 | axon terminus | - | IEA | Component |
| GO:0046686 | response to cadmium ion | - | IEA | Process |
| GO:0047801 | L-cysteine:2-oxoglutarate aminotransferase activity | - | ISS | Function |
| GO:0051384 | response to glucocorticoid | 8396422. | IEP | Process |
| GO:0051481 | negative regulation of cytosolic calcium ion concentration | - | IEA | Process |
| GO:0051902 | negative regulation of mitochondrial depolarization | - | IEA | Process |
| GO:0055089 | fatty acid homeostasis | - | IEA | Process |
| GO:0060290 | transdifferentiation | - | IEA | Process |
| GO:0070062 | extracellular exosome | 19056867.19199708.20458337.23533145. | HDA | Component |
| GO:0071260 | cellular response to mechanical stimulus | - | IEA | Process |
| GO:1990267 | response to transition metal nanoparticle | - | IEA | Process |