
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| PAAD | |||
| SARC |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| SARC | ||
| MESO | ||
| SKCM | ||
| PRAD | ||
| KIRP | ||
| PAAD | ||
| PCPG | ||
| READ | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LUAD | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000455200 | NCAPH-205 | 2952 | - | ENSP00000407308 | 730 (aa) | - | E9PHA2 |
| ENST00000435349 | NCAPH-203 | 779 | - | ENSP00000415162 | 182 (aa) | - | H7C415 |
| ENST00000240423 | NCAPH-201 | 2763 | XM_005263908 | ENSP00000240423 | 741 (aa) | XP_005263965 | Q15003 |
| ENST00000477409 | NCAPH-207 | 1573 | - | - | - (aa) | - | - |
| ENST00000427946 | NCAPH-202 | 2363 | - | ENSP00000400774 | 605 (aa) | - | Q15003 |
| ENST00000435975 | NCAPH-204 | 1852 | - | ENSP00000405237 | 607 (aa) | - | C9J470 |
| ENST00000456906 | NCAPH-206 | 673 | - | ENSP00000401227 | 216 (aa) | - | C9JZP1 |
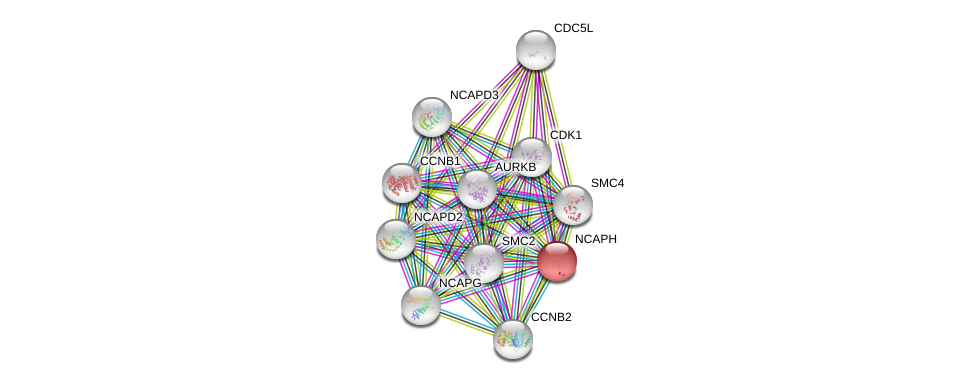
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000796 | condensin complex | 11136719. | IDA | Component |
| GO:0000799 | nuclear condensin complex | 21873635. | IBA | Component |
| GO:0003682 | chromatin binding | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 17268547.21633354.21730191.26496610. | IPI | Function |
| GO:0005634 | nucleus | - | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0007076 | mitotic chromosome condensation | 21873635. | IBA | Process |
| GO:0007076 | mitotic chromosome condensation | 11136719.11694586. | IDA | Process |
| GO:0007076 | mitotic chromosome condensation | 27737959. | IMP | Process |
| GO:0010032 | meiotic chromosome condensation | - | IEA | Process |
| GO:0016020 | membrane | 19946888. | HDA | Component |
| GO:0044547 | DNA topoisomerase binding | 21873635. | IBA | Function |
| GO:0051301 | cell division | - | IEA | Process |
| GO:0051309 | female meiosis chromosome separation | - | IEA | Process |
| GO:0072587 | DNA topoisomerase (ATP-hydrolyzing) activator activity | 21873635. | IBA | Function |
| GO:2000373 | positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity | - | IEA | Process |