
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 15033445 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| LAML | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRC | |||
| MESO | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| SARC | ||
| MESO | ||
| UCS | ||
| HNSC | ||
| KIRP | ||
| LAML | ||
| LUAD | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000241463 | MMR_HSR1 | PF01926.23 | 0.00035 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000475647 | RASL11A-202 | 415 | - | - | - (aa) | - | - |
| ENST00000480803 | RASL11A-203 | 507 | - | - | - (aa) | - | - |
| ENST00000241463 | RASL11A-201 | 1543 | XM_024449347 | ENSP00000241463 | 242 (aa) | XP_024305115 | Q6T310 |
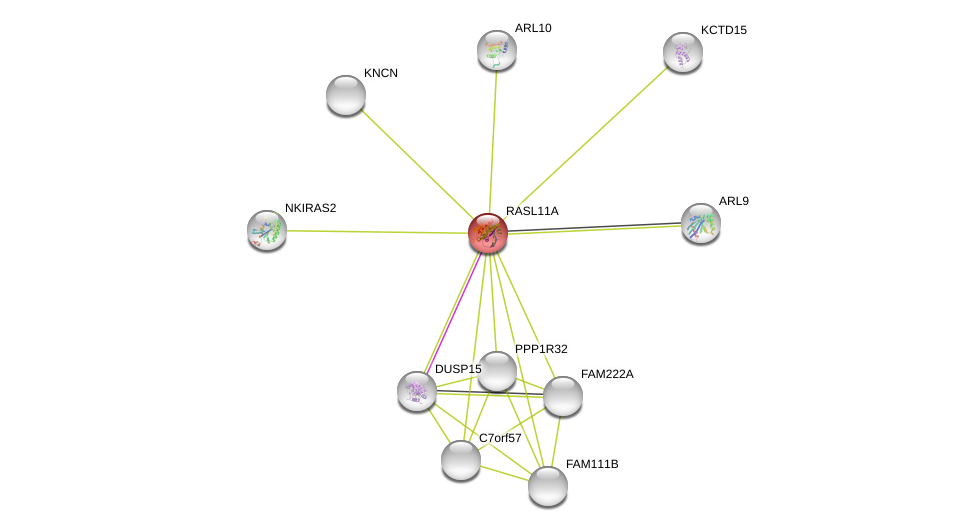
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000122035 | RASL11A | 99 | 92.500 | ENSAMEG00000015812 | RASL11A | 99 | 92.500 | Ailuropoda_melanoleuca |
| ENSG00000122035 | RASL11A | 98 | 72.996 | ENSACAG00000003190 | RASL11A | 100 | 72.996 | Anolis_carolinensis |
| ENSG00000122035 | RASL11A | 100 | 96.694 | ENSANAG00000027999 | RASL11A | 100 | 96.694 | Aotus_nancymaae |
| ENSG00000122035 | RASL11A | 100 | 96.694 | ENSCJAG00000019369 | RASL11A | 100 | 96.694 | Callithrix_jacchus |
| ENSG00000122035 | RASL11A | 100 | 93.388 | ENSCAFG00000006798 | RASL11A | 100 | 93.388 | Canis_familiaris |
| ENSG00000122035 | RASL11A | 100 | 93.388 | ENSCAFG00020025312 | RASL11A | 100 | 93.388 | Canis_lupus_dingo |
| ENSG00000122035 | RASL11A | 100 | 90.083 | ENSCPOG00000031264 | RASL11A | 100 | 90.083 | Cavia_porcellus |
| ENSG00000122035 | RASL11A | 100 | 97.107 | ENSCCAG00000021842 | RASL11A | 100 | 97.107 | Cebus_capucinus |
| ENSG00000122035 | RASL11A | 100 | 99.174 | ENSCATG00000033838 | RASL11A | 100 | 99.174 | Cercocebus_atys |
| ENSG00000122035 | RASL11A | 100 | 98.760 | ENSCSAG00000017985 | RASL11A | 100 | 98.760 | Chlorocebus_sabaeus |
| ENSG00000122035 | RASL11A | 98 | 78.481 | ENSCPBG00000020480 | RASL11A | 52 | 78.481 | Chrysemys_picta_bellii |
| ENSG00000122035 | RASL11A | 100 | 98.347 | ENSCANG00000038972 | RASL11A | 100 | 98.347 | Colobus_angolensis_palliatus |
| ENSG00000122035 | RASL11A | 100 | 91.736 | ENSCGRG00001014752 | Rasl11a | 100 | 91.736 | Cricetulus_griseus_chok1gshd |
| ENSG00000122035 | RASL11A | 100 | 91.736 | ENSCGRG00000013361 | Rasl11a | 100 | 91.736 | Cricetulus_griseus_crigri |
| ENSG00000122035 | RASL11A | 100 | 90.083 | ENSDORG00000026016 | Rasl11a | 100 | 90.083 | Dipodomys_ordii |
| ENSG00000122035 | RASL11A | 100 | 92.149 | ENSEASG00005022497 | RASL11A | 100 | 92.149 | Equus_asinus_asinus |
| ENSG00000122035 | RASL11A | 100 | 92.562 | ENSECAG00000012858 | RASL11A | 100 | 92.562 | Equus_caballus |
| ENSG00000122035 | RASL11A | 100 | 90.496 | ENSFCAG00000006749 | RASL11A | 100 | 90.496 | Felis_catus |
| ENSG00000122035 | RASL11A | 98 | 71.548 | ENSFALG00000008621 | RASL11A | 100 | 71.548 | Ficedula_albicollis |
| ENSG00000122035 | RASL11A | 100 | 90.496 | ENSFDAG00000008237 | RASL11A | 100 | 90.496 | Fukomys_damarensis |
| ENSG00000122035 | RASL11A | 98 | 66.387 | ENSGALG00000017099 | RASL11A | 100 | 66.387 | Gallus_gallus |
| ENSG00000122035 | RASL11A | 98 | 78.059 | ENSGAGG00000015067 | RASL11A | 100 | 78.059 | Gopherus_agassizii |
| ENSG00000122035 | RASL11A | 100 | 99.587 | ENSGGOG00000008785 | RASL11A | 100 | 99.587 | Gorilla_gorilla |
| ENSG00000122035 | RASL11A | 98 | 91.176 | ENSHGLG00000005304 | RASL11A | 98 | 91.176 | Heterocephalus_glaber_female |
| ENSG00000122035 | RASL11A | 98 | 91.176 | ENSHGLG00100011809 | RASL11A | 98 | 91.176 | Heterocephalus_glaber_male |
| ENSG00000122035 | RASL11A | 100 | 94.215 | ENSSTOG00000023879 | RASL11A | 100 | 94.215 | Ictidomys_tridecemlineatus |
| ENSG00000122035 | RASL11A | 99 | 89.583 | ENSLAFG00000003673 | RASL11A | 98 | 89.583 | Loxodonta_africana |
| ENSG00000122035 | RASL11A | 100 | 99.174 | ENSMFAG00000044628 | RASL11A | 100 | 99.174 | Macaca_fascicularis |
| ENSG00000122035 | RASL11A | 100 | 98.760 | ENSMNEG00000033345 | RASL11A | 100 | 98.760 | Macaca_nemestrina |
| ENSG00000122035 | RASL11A | 100 | 99.174 | ENSMLEG00000036089 | RASL11A | 100 | 99.174 | Mandrillus_leucophaeus |
| ENSG00000122035 | RASL11A | 100 | 91.322 | ENSMAUG00000019356 | Rasl11a | 100 | 91.322 | Mesocricetus_auratus |
| ENSG00000122035 | RASL11A | 100 | 93.802 | ENSMICG00000029724 | RASL11A | 100 | 93.802 | Microcebus_murinus |
| ENSG00000122035 | RASL11A | 100 | 92.562 | ENSMOCG00000000626 | Rasl11a | 100 | 92.562 | Microtus_ochrogaster |
| ENSG00000122035 | RASL11A | 100 | 83.471 | ENSMODG00000008941 | RASL11A | 100 | 83.471 | Monodelphis_domestica |
| ENSG00000122035 | RASL11A | 100 | 90.083 | MGP_CAROLIEiJ_G0028030 | Rasl11a | 100 | 90.083 | Mus_caroli |
| ENSG00000122035 | RASL11A | 100 | 89.256 | ENSMUSG00000029641 | Rasl11a | 100 | 89.256 | Mus_musculus |
| ENSG00000122035 | RASL11A | 100 | 90.496 | MGP_PahariEiJ_G0024596 | Rasl11a | 100 | 90.496 | Mus_pahari |
| ENSG00000122035 | RASL11A | 100 | 90.083 | MGP_SPRETEiJ_G0029028 | Rasl11a | 100 | 90.083 | Mus_spretus |
| ENSG00000122035 | RASL11A | 100 | 92.975 | ENSMPUG00000009362 | RASL11A | 100 | 92.975 | Mustela_putorius_furo |
| ENSG00000122035 | RASL11A | 100 | 94.215 | ENSMLUG00000016335 | RASL11A | 100 | 94.215 | Myotis_lucifugus |
| ENSG00000122035 | RASL11A | 100 | 92.562 | ENSNGAG00000021721 | Rasl11a | 100 | 92.562 | Nannospalax_galili |
| ENSG00000122035 | RASL11A | 100 | 100.000 | ENSNLEG00000000329 | RASL11A | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000122035 | RASL11A | 100 | 90.496 | ENSODEG00000005775 | RASL11A | 100 | 90.496 | Octodon_degus |
| ENSG00000122035 | RASL11A | 100 | 91.322 | ENSOCUG00000008121 | RASL11A | 100 | 91.322 | Oryctolagus_cuniculus |
| ENSG00000122035 | RASL11A | 98 | 93.697 | ENSOGAG00000008177 | RASL11A | 98 | 93.697 | Otolemur_garnettii |
| ENSG00000122035 | RASL11A | 100 | 100.000 | ENSPPAG00000027996 | RASL11A | 100 | 100.000 | Pan_paniscus |
| ENSG00000122035 | RASL11A | 100 | 100.000 | ENSPTRG00000005729 | RASL11A | 100 | 100.000 | Pan_troglodytes |
| ENSG00000122035 | RASL11A | 100 | 98.760 | ENSPANG00000004651 | RASL11A | 100 | 98.760 | Papio_anubis |
| ENSG00000122035 | RASL11A | 98 | 78.571 | ENSPSIG00000011613 | RASL11A | 100 | 78.571 | Pelodiscus_sinensis |
| ENSG00000122035 | RASL11A | 100 | 92.149 | ENSPEMG00000004475 | Rasl11a | 100 | 92.149 | Peromyscus_maniculatus_bairdii |
| ENSG00000122035 | RASL11A | 100 | 100.000 | ENSPPYG00000005234 | RASL11A | 100 | 100.000 | Pongo_abelii |
| ENSG00000122035 | RASL11A | 100 | 94.215 | ENSPCOG00000009447 | RASL11A | 100 | 94.215 | Propithecus_coquereli |
| ENSG00000122035 | RASL11A | 100 | 91.322 | ENSRNOG00000000956 | Rasl11a | 100 | 91.322 | Rattus_norvegicus |
| ENSG00000122035 | RASL11A | 100 | 98.347 | ENSRBIG00000043749 | RASL11A | 100 | 98.347 | Rhinopithecus_bieti |
| ENSG00000122035 | RASL11A | 100 | 98.347 | ENSRROG00000032810 | RASL11A | 100 | 98.347 | Rhinopithecus_roxellana |
| ENSG00000122035 | RASL11A | 100 | 96.694 | ENSSBOG00000025363 | RASL11A | 100 | 96.694 | Saimiri_boliviensis_boliviensis |
| ENSG00000122035 | RASL11A | 100 | 82.231 | ENSSHAG00000016468 | RASL11A | 100 | 82.231 | Sarcophilus_harrisii |
| ENSG00000122035 | RASL11A | 100 | 88.066 | ENSSARG00000011269 | RASL11A | 100 | 88.066 | Sorex_araneus |
| ENSG00000122035 | RASL11A | 99 | 77.917 | ENSSPUG00000005315 | RASL11A | 100 | 77.917 | Sphenodon_punctatus |
| ENSG00000122035 | RASL11A | 100 | 94.215 | ENSUAMG00000016809 | RASL11A | 100 | 94.215 | Ursus_americanus |
| ENSG00000122035 | RASL11A | 100 | 93.388 | ENSVVUG00000009191 | RASL11A | 100 | 93.388 | Vulpes_vulpes |
| ENSG00000122035 | RASL11A | 99 | 68.313 | ENSXETG00000030983 | RASL11A | 97 | 68.313 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003924 | GTPase activity | - | IEA | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005730 | nucleolus | - | ISS | Component |
| GO:0007165 | signal transduction | - | IEA | Process |
| GO:0016020 | membrane | - | IEA | Component |
| GO:0045943 | positive regulation of transcription by RNA polymerase I | - | ISS | Process |