
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRC | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| STAD | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| LUSC | ||
| KIRP | ||
| PAAD | ||
| PCPG | ||
| UCEC | ||
| LGG | ||
| LUAD |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000243298 | MMR_HSR1 | PF01926.23 | 2e-05 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000243298 | RAB9B-201 | 3748 | - | ENSP00000243298 | 201 (aa) | - | Q9NP90 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa05162 | Measles | KEGG |
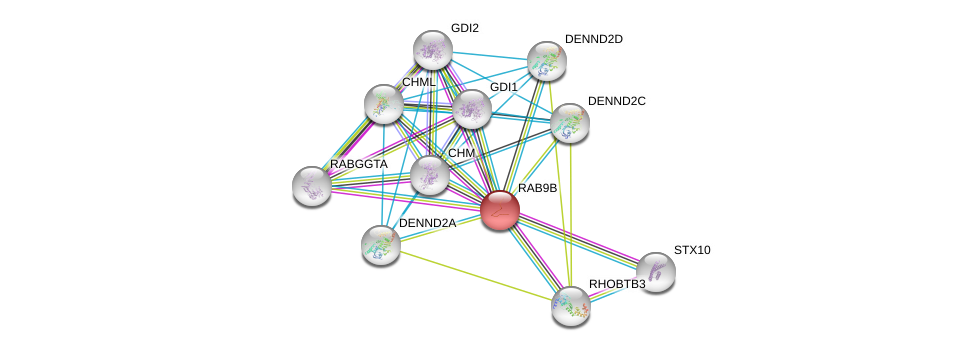
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000123570 | RAB9B | 100 | 76.238 | ENSAPOG00000007269 | rab9b | 89 | 76.238 | Acanthochromis_polyacanthus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSAMEG00000018531 | RAB9B | 100 | 99.502 | Ailuropoda_melanoleuca |
| ENSG00000123570 | RAB9B | 100 | 77.228 | ENSACIG00000007739 | rab9b | 99 | 77.228 | Amphilophus_citrinellus |
| ENSG00000123570 | RAB9B | 100 | 76.238 | ENSAOCG00000022810 | rab9b | 100 | 76.238 | Amphiprion_ocellaris |
| ENSG00000123570 | RAB9B | 100 | 76.238 | ENSAPEG00000014149 | rab9b | 100 | 76.238 | Amphiprion_percula |
| ENSG00000123570 | RAB9B | 100 | 77.723 | ENSATEG00000004488 | rab9b | 96 | 77.723 | Anabas_testudineus |
| ENSG00000123570 | RAB9B | 100 | 93.532 | ENSAPLG00000001949 | RAB9B | 100 | 93.532 | Anas_platyrhynchos |
| ENSG00000123570 | RAB9B | 100 | 86.070 | ENSACAG00000014318 | RAB9B | 100 | 86.070 | Anolis_carolinensis |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSANAG00000031345 | RAB9B | 100 | 99.502 | Aotus_nancymaae |
| ENSG00000123570 | RAB9B | 100 | 76.733 | ENSACLG00000004795 | rab9b | 91 | 76.733 | Astatotilapia_calliptera |
| ENSG00000123570 | RAB9B | 100 | 79.703 | ENSAMXG00000026476 | rab9b | 90 | 79.703 | Astyanax_mexicanus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSBTAG00000019820 | RAB9B | 100 | 99.502 | Bos_taurus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSCJAG00000003347 | RAB9B | 100 | 99.502 | Callithrix_jacchus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSCAFG00000017812 | RAB9B | 100 | 99.502 | Canis_familiaris |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSCHIG00000002776 | RAB9B | 100 | 99.502 | Capra_hircus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSTSYG00000008912 | RAB9B | 100 | 99.502 | Carlito_syrichta |
| ENSG00000123570 | RAB9B | 100 | 99.005 | ENSCCAG00000017812 | RAB9B | 100 | 99.005 | Cebus_capucinus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSCSAG00000000315 | RAB9B | 100 | 99.502 | Chlorocebus_sabaeus |
| ENSG00000123570 | RAB9B | 100 | 98.507 | ENSCHOG00000012678 | RAB9B | 100 | 98.507 | Choloepus_hoffmanni |
| ENSG00000123570 | RAB9B | 100 | 93.532 | ENSCPBG00000001542 | RAB9B | 100 | 93.532 | Chrysemys_picta_bellii |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSCANG00000012854 | RAB9B | 100 | 99.502 | Colobus_angolensis_palliatus |
| ENSG00000123570 | RAB9B | 100 | 98.010 | ENSCGRG00001001220 | Rab9b | 100 | 98.010 | Cricetulus_griseus_chok1gshd |
| ENSG00000123570 | RAB9B | 100 | 97.015 | ENSCGRG00000002809 | Rab9b | 100 | 97.015 | Cricetulus_griseus_crigri |
| ENSG00000123570 | RAB9B | 100 | 78.218 | ENSCSEG00000014467 | rab9b | 92 | 78.218 | Cynoglossus_semilaevis |
| ENSG00000123570 | RAB9B | 100 | 75.743 | ENSCVAG00000021826 | rab9b | 100 | 75.743 | Cyprinodon_variegatus |
| ENSG00000123570 | RAB9B | 100 | 77.228 | ENSDARG00000103391 | rab9b | 100 | 77.228 | Danio_rerio |
| ENSG00000123570 | RAB9B | 100 | 99.005 | ENSDNOG00000032358 | RAB9B | 100 | 99.005 | Dasypus_novemcinctus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSETEG00000018698 | RAB9B | 100 | 99.502 | Echinops_telfairi |
| ENSG00000123570 | RAB9B | 86 | 68.786 | ENSEBUG00000004967 | rab9b | 84 | 68.786 | Eptatretus_burgeri |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSEASG00005002563 | RAB9B | 100 | 99.502 | Equus_asinus_asinus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSECAG00000022069 | RAB9B | 100 | 99.502 | Equus_caballus |
| ENSG00000123570 | RAB9B | 100 | 99.005 | ENSEEUG00000005264 | RAB9B | 100 | 99.005 | Erinaceus_europaeus |
| ENSG00000123570 | RAB9B | 100 | 75.862 | ENSELUG00000004378 | rab9b | 100 | 75.862 | Esox_lucius |
| ENSG00000123570 | RAB9B | 100 | 100.000 | ENSFCAG00000000424 | RAB9B | 100 | 100.000 | Felis_catus |
| ENSG00000123570 | RAB9B | 100 | 95.025 | ENSFALG00000015153 | RAB9B | 100 | 95.025 | Ficedula_albicollis |
| ENSG00000123570 | RAB9B | 100 | 99.005 | ENSFDAG00000014776 | RAB9B | 100 | 99.005 | Fukomys_damarensis |
| ENSG00000123570 | RAB9B | 100 | 75.248 | ENSFHEG00000009417 | rab9b | 92 | 75.248 | Fundulus_heteroclitus |
| ENSG00000123570 | RAB9B | 100 | 79.310 | ENSGMOG00000020194 | rab9b | 99 | 79.310 | Gadus_morhua |
| ENSG00000123570 | RAB9B | 100 | 93.532 | ENSGALG00000004918 | RAB9B | 100 | 93.532 | Gallus_gallus |
| ENSG00000123570 | RAB9B | 100 | 75.248 | ENSGAFG00000004099 | rab9b | 90 | 75.248 | Gambusia_affinis |
| ENSG00000123570 | RAB9B | 100 | 76.733 | ENSGACG00000018291 | rab9b | 99 | 76.733 | Gasterosteus_aculeatus |
| ENSG00000123570 | RAB9B | 100 | 92.537 | ENSGAGG00000012314 | RAB9B | 100 | 92.537 | Gopherus_agassizii |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSGGOG00000009507 | RAB9B | 100 | 99.502 | Gorilla_gorilla |
| ENSG00000123570 | RAB9B | 100 | 76.733 | ENSHBUG00000001595 | rab9b | 91 | 76.733 | Haplochromis_burtoni |
| ENSG00000123570 | RAB9B | 100 | 98.010 | ENSHGLG00000000448 | RAB9B | 100 | 98.010 | Heterocephalus_glaber_female |
| ENSG00000123570 | RAB9B | 100 | 98.010 | ENSHGLG00100003190 | RAB9B | 100 | 98.010 | Heterocephalus_glaber_male |
| ENSG00000123570 | RAB9B | 100 | 78.109 | ENSHCOG00000005024 | rab9b | 99 | 78.109 | Hippocampus_comes |
| ENSG00000123570 | RAB9B | 100 | 70.297 | ENSIPUG00000016561 | rab9b | 91 | 70.297 | Ictalurus_punctatus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSSTOG00000004081 | RAB9B | 100 | 99.502 | Ictidomys_tridecemlineatus |
| ENSG00000123570 | RAB9B | 100 | 76.238 | ENSKMAG00000019964 | rab9b | 100 | 76.238 | Kryptolebias_marmoratus |
| ENSG00000123570 | RAB9B | 99 | 77.500 | ENSLBEG00000011478 | rab9b | 92 | 77.500 | Labrus_bergylta |
| ENSG00000123570 | RAB9B | 89 | 89.326 | ENSLACG00000017891 | RAB9B | 84 | 89.326 | Latimeria_chalumnae |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSLAFG00000000166 | RAB9B | 100 | 99.502 | Loxodonta_africana |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSMFAG00000010320 | RAB9B | 100 | 99.502 | Macaca_fascicularis |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSMMUG00000021198 | RAB9B | 100 | 99.502 | Macaca_mulatta |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSMLEG00000027985 | RAB9B | 100 | 99.502 | Mandrillus_leucophaeus |
| ENSG00000123570 | RAB9B | 100 | 77.073 | ENSMAMG00000009927 | rab9b | 100 | 77.073 | Mastacembelus_armatus |
| ENSG00000123570 | RAB9B | 100 | 76.733 | ENSMZEG00005010392 | rab9b | 91 | 76.733 | Maylandia_zebra |
| ENSG00000123570 | RAB9B | 100 | 98.507 | ENSMAUG00000010219 | Rab9b | 100 | 98.507 | Mesocricetus_auratus |
| ENSG00000123570 | RAB9B | 100 | 99.005 | ENSMICG00000003057 | RAB9B | 100 | 99.005 | Microcebus_murinus |
| ENSG00000123570 | RAB9B | 100 | 96.517 | ENSMOCG00000010381 | Rab9b | 100 | 96.517 | Microtus_ochrogaster |
| ENSG00000123570 | RAB9B | 99 | 77.612 | ENSMMOG00000008669 | rab9b | 91 | 77.612 | Mola_mola |
| ENSG00000123570 | RAB9B | 100 | 95.025 | ENSMODG00000011531 | RAB9B | 100 | 95.025 | Monodelphis_domestica |
| ENSG00000123570 | RAB9B | 100 | 77.228 | ENSMALG00000007664 | rab9b | 94 | 77.228 | Monopterus_albus |
| ENSG00000123570 | RAB9B | 100 | 98.010 | MGP_CAROLIEiJ_G0033427 | Rab9b | 100 | 98.010 | Mus_caroli |
| ENSG00000123570 | RAB9B | 100 | 98.010 | ENSMUSG00000043463 | Rab9b | 100 | 98.010 | Mus_musculus |
| ENSG00000123570 | RAB9B | 100 | 98.010 | MGP_PahariEiJ_G0031967 | Rab9b | 100 | 98.010 | Mus_pahari |
| ENSG00000123570 | RAB9B | 100 | 98.010 | MGP_SPRETEiJ_G0034594 | Rab9b | 100 | 98.010 | Mus_spretus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSMPUG00000019218 | RAB9B | 100 | 99.502 | Mustela_putorius_furo |
| ENSG00000123570 | RAB9B | 100 | 99.005 | ENSMLUG00000028856 | RAB9B | 100 | 99.005 | Myotis_lucifugus |
| ENSG00000123570 | RAB9B | 100 | 98.010 | ENSNGAG00000011436 | Rab9b | 100 | 98.010 | Nannospalax_galili |
| ENSG00000123570 | RAB9B | 100 | 76.733 | ENSNBRG00000013940 | rab9b | 91 | 76.733 | Neolamprologus_brichardi |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSNLEG00000018324 | RAB9B | 100 | 99.502 | Nomascus_leucogenys |
| ENSG00000123570 | RAB9B | 100 | 94.527 | ENSMEUG00000000993 | RAB9B | 100 | 94.527 | Notamacropus_eugenii |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSOPRG00000015421 | RAB9B | 100 | 99.502 | Ochotona_princeps |
| ENSG00000123570 | RAB9B | 100 | 76.733 | ENSONIG00000020581 | rab9b | 100 | 76.733 | Oreochromis_niloticus |
| ENSG00000123570 | RAB9B | 100 | 95.025 | ENSOANG00000004230 | RAB9B | 100 | 95.025 | Ornithorhynchus_anatinus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSOCUG00000014809 | RAB9B | 100 | 99.502 | Oryctolagus_cuniculus |
| ENSG00000123570 | RAB9B | 100 | 75.248 | ENSORLG00000006405 | rab9b | 96 | 75.248 | Oryzias_latipes |
| ENSG00000123570 | RAB9B | 100 | 76.238 | ENSORLG00020009158 | rab9b | 96 | 76.238 | Oryzias_latipes_hni |
| ENSG00000123570 | RAB9B | 100 | 75.248 | ENSORLG00015004265 | rab9b | 97 | 75.248 | Oryzias_latipes_hsok |
| ENSG00000123570 | RAB9B | 100 | 76.733 | ENSOMEG00000008327 | rab9b | 93 | 76.733 | Oryzias_melastigma |
| ENSG00000123570 | RAB9B | 100 | 99.005 | ENSOGAG00000015082 | RAB9B | 100 | 99.005 | Otolemur_garnettii |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSOARG00000009986 | RAB9B | 100 | 99.502 | Ovis_aries |
| ENSG00000123570 | RAB9B | 100 | 100.000 | ENSPPAG00000002892 | RAB9B | 100 | 100.000 | Pan_paniscus |
| ENSG00000123570 | RAB9B | 100 | 100.000 | ENSPPRG00000011181 | RAB9B | 100 | 100.000 | Panthera_pardus |
| ENSG00000123570 | RAB9B | 100 | 100.000 | ENSPTIG00000009359 | RAB9B | 100 | 100.000 | Panthera_tigris_altaica |
| ENSG00000123570 | RAB9B | 100 | 100.000 | ENSPTRG00000028227 | RAB9B | 100 | 100.000 | Pan_troglodytes |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSPANG00000018955 | RAB9B | 100 | 99.502 | Papio_anubis |
| ENSG00000123570 | RAB9B | 100 | 78.818 | ENSPKIG00000014623 | rab9b | 92 | 78.818 | Paramormyrops_kingsleyae |
| ENSG00000123570 | RAB9B | 100 | 93.532 | ENSPSIG00000001871 | RAB9B | 100 | 93.532 | Pelodiscus_sinensis |
| ENSG00000123570 | RAB9B | 100 | 77.228 | ENSPMGG00000002496 | rab9b | 100 | 77.228 | Periophthalmus_magnuspinnatus |
| ENSG00000123570 | RAB9B | 100 | 95.025 | ENSPCIG00000020415 | RAB9B | 100 | 95.025 | Phascolarctos_cinereus |
| ENSG00000123570 | RAB9B | 100 | 75.248 | ENSPFOG00000020060 | rab9b | 100 | 75.248 | Poecilia_formosa |
| ENSG00000123570 | RAB9B | 100 | 75.248 | ENSPLAG00000011866 | rab9b | 91 | 75.248 | Poecilia_latipinna |
| ENSG00000123570 | RAB9B | 100 | 75.248 | ENSPMEG00000024399 | rab9b | 91 | 75.248 | Poecilia_mexicana |
| ENSG00000123570 | RAB9B | 100 | 75.248 | ENSPMEG00000013734 | rab9b | 91 | 75.248 | Poecilia_mexicana |
| ENSG00000123570 | RAB9B | 100 | 74.752 | ENSPREG00000006970 | rab9b | 92 | 74.752 | Poecilia_reticulata |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSPPYG00000020591 | RAB9B | 100 | 99.502 | Pongo_abelii |
| ENSG00000123570 | RAB9B | 100 | 76.119 | ENSPCAG00000013418 | RAB9B | 100 | 76.119 | Procavia_capensis |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSPCOG00000002853 | RAB9B | 100 | 99.502 | Propithecus_coquereli |
| ENSG00000123570 | RAB9B | 55 | 100.000 | ENSPVAG00000008773 | RAB9B | 100 | 100.000 | Pteropus_vampyrus |
| ENSG00000123570 | RAB9B | 100 | 76.733 | ENSPNYG00000000233 | rab9b | 91 | 76.733 | Pundamilia_nyererei |
| ENSG00000123570 | RAB9B | 100 | 79.208 | ENSPNAG00000018132 | rab9b | 90 | 79.208 | Pygocentrus_nattereri |
| ENSG00000123570 | RAB9B | 100 | 98.507 | ENSRNOG00000047960 | Rab9b | 100 | 98.507 | Rattus_norvegicus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSRROG00000025642 | RAB9B | 100 | 99.502 | Rhinopithecus_roxellana |
| ENSG00000123570 | RAB9B | 100 | 94.527 | ENSSHAG00000012788 | RAB9B | 100 | 94.527 | Sarcophilus_harrisii |
| ENSG00000123570 | RAB9B | 100 | 79.310 | ENSSFOG00015010492 | rab9b | 100 | 79.310 | Scleropages_formosus |
| ENSG00000123570 | RAB9B | 100 | 77.228 | ENSSMAG00000006709 | rab9b | 98 | 77.228 | Scophthalmus_maximus |
| ENSG00000123570 | RAB9B | 100 | 78.218 | ENSSDUG00000007575 | rab9b | 99 | 78.218 | Seriola_dumerili |
| ENSG00000123570 | RAB9B | 100 | 78.218 | ENSSLDG00000009351 | rab9b | 99 | 78.218 | Seriola_lalandi_dorsalis |
| ENSG00000123570 | RAB9B | 100 | 98.507 | ENSSARG00000006891 | RAB9B | 100 | 98.507 | Sorex_araneus |
| ENSG00000123570 | RAB9B | 100 | 94.030 | ENSSPUG00000017531 | RAB9B | 100 | 94.030 | Sphenodon_punctatus |
| ENSG00000123570 | RAB9B | 100 | 75.743 | ENSSPAG00000009806 | rab9b | 92 | 75.743 | Stegastes_partitus |
| ENSG00000123570 | RAB9B | 100 | 99.005 | ENSSSCG00000032672 | RAB9B | 100 | 99.005 | Sus_scrofa |
| ENSG00000123570 | RAB9B | 100 | 95.025 | ENSTGUG00000006516 | RAB9B | 100 | 95.025 | Taeniopygia_guttata |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSTBEG00000006468 | RAB9B | 100 | 99.502 | Tupaia_belangeri |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSTTRG00000003956 | RAB9B | 100 | 99.502 | Tursiops_truncatus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSUAMG00000018451 | RAB9B | 100 | 99.502 | Ursus_americanus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSUMAG00000001213 | RAB9B | 100 | 99.502 | Ursus_maritimus |
| ENSG00000123570 | RAB9B | 100 | 99.502 | ENSVVUG00000001381 | RAB9B | 100 | 99.502 | Vulpes_vulpes |
| ENSG00000123570 | RAB9B | 100 | 85.075 | ENSXETG00000018133 | rab9b | 100 | 85.075 | Xenopus_tropicalis |
| ENSG00000123570 | RAB9B | 100 | 75.248 | ENSXCOG00000009094 | rab9b | 100 | 75.248 | Xiphophorus_couchianus |
| ENSG00000123570 | RAB9B | 98 | 76.263 | ENSXMAG00000019499 | rab9b | 96 | 76.263 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003924 | GTPase activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 19490898.20048159. | IPI | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005829 | cytosol | - | IEA | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0006886 | intracellular protein transport | 21873635. | IBA | Process |
| GO:0019003 | GDP binding | 20937701. | IDA | Function |
| GO:0030667 | secretory granule membrane | - | TAS | Component |
| GO:0030670 | phagocytic vesicle membrane | - | IEA | Component |
| GO:0032482 | Rab protein signal transduction | 21873635. | IBA | Process |
| GO:0042147 | retrograde transport, endosome to Golgi | 21873635. | IBA | Process |
| GO:0042802 | identical protein binding | - | IEA | Function |
| GO:0043312 | neutrophil degranulation | - | TAS | Process |
| GO:0045335 | phagocytic vesicle | 21255211. | IDA | Component |