
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| PAAD | |||
| PAAD |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| ACC | ||
| ESCA | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| BLCA | ||
| KICH | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| THCA | ||
| LUAD | ||
| UVM | ||
| OV |
| PID | Title | Method | Time | Author | Doi |
|---|---|---|---|---|---|
| 27453046 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000462821 | METTL8-211 | 623 | - | - | - (aa) | - | - |
| ENST00000392599 | METTL8-202 | 828 | - | ENSP00000376377 | 226 (aa) | - | B5ME25 |
| ENST00000638989 | METTL8-218 | 876 | - | ENSP00000492427 | 291 (aa) | - | Q9H825 |
| ENST00000438609 | METTL8-204 | 981 | - | ENSP00000407180 | 215 (aa) | - | H7C2P8 |
| ENST00000442541 | METTL8-205 | 563 | - | ENSP00000411942 | 83 (aa) | - | C9J3F1 |
| ENST00000483284 | METTL8-216 | 1880 | - | - | - (aa) | - | - |
| ENST00000375258 | METTL8-201 | 8203 | - | ENSP00000364407 | 379 (aa) | - | E7ETE0 |
| ENST00000447486 | METTL8-207 | 2246 | - | ENSP00000409025 | 241 (aa) | - | Q9H825 |
| ENST00000477130 | METTL8-215 | 445 | - | - | - (aa) | - | - |
| ENST00000464491 | METTL8-213 | 744 | - | - | - (aa) | - | - |
| ENST00000460188 | METTL8-209 | 622 | NR_135568 | - | - (aa) | - | - |
| ENST00000470773 | METTL8-214 | 575 | - | - | - (aa) | - | - |
| ENST00000640734 | METTL8-219 | 726 | - | ENSP00000491656 | 241 (aa) | - | Q9H825 |
| ENST00000463392 | METTL8-212 | 697 | - | - | - (aa) | - | - |
| ENST00000442778 | METTL8-206 | 578 | - | ENSP00000404646 | 153 (aa) | - | C9JE69 |
| ENST00000460539 | METTL8-210 | 677 | - | - | - (aa) | - | - |
| ENST00000453846 | METTL8-208 | 558 | - | ENSP00000411589 | 105 (aa) | - | C9J6U8 |
| ENST00000612742 | METTL8-217 | 8132 | XM_011511875 | ENSP00000480056 | 407 (aa) | XP_011510177 | B3KW44 |
| ENST00000392604 | METTL8-203 | 1697 | - | ENSP00000376382 | 291 (aa) | - | Q9H825 |
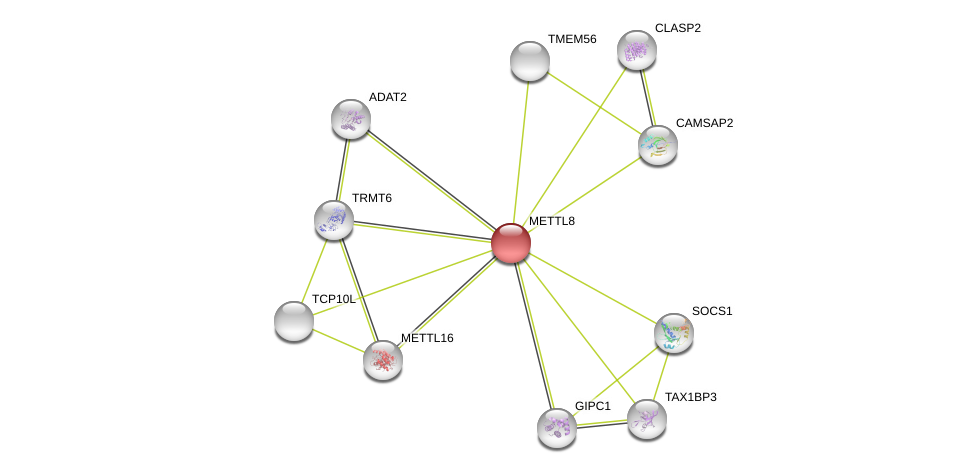
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0005634 | nucleus | - | IEA | Component |
| GO:0005737 | cytoplasm | - | IEA | Component |
| GO:0030488 | tRNA methylation | - | IEA | Process |
| GO:0052735 | tRNA (cytosine-3-)-methyltransferase activity | 21873635. | IBA | Function |