
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KICH | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| CESC | |||
| HNSC | |||
| LIHC | |||
| LUSC | |||
| PAAD | |||
| PRAD | |||
| SARC | |||
| SKCM | |||
| UVM |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| MESO | ||
| HNSC | ||
| SKCM | ||
| KIRP | ||
| READ | ||
| LAML | ||
| GBM | ||
| LGG | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000369270 | zf-C2H2 | PF00096.26 | 5.3e-40 | 1 | 4 |
| ENSP00000369270 | zf-C2H2 | PF00096.26 | 5.3e-40 | 2 | 4 |
| ENSP00000369270 | zf-C2H2 | PF00096.26 | 5.3e-40 | 3 | 4 |
| ENSP00000369270 | zf-C2H2 | PF00096.26 | 5.3e-40 | 4 | 4 |
| ENSP00000305560 | zf-C2H2 | PF00096.26 | 9.5e-39 | 1 | 4 |
| ENSP00000305560 | zf-C2H2 | PF00096.26 | 9.5e-39 | 2 | 4 |
| ENSP00000305560 | zf-C2H2 | PF00096.26 | 9.5e-39 | 3 | 4 |
| ENSP00000305560 | zf-C2H2 | PF00096.26 | 9.5e-39 | 4 | 4 |
| ENSP00000369265 | zf-C2H2 | PF00096.26 | 9.5e-39 | 1 | 4 |
| ENSP00000369265 | zf-C2H2 | PF00096.26 | 9.5e-39 | 2 | 4 |
| ENSP00000369265 | zf-C2H2 | PF00096.26 | 9.5e-39 | 3 | 4 |
| ENSP00000369265 | zf-C2H2 | PF00096.26 | 9.5e-39 | 4 | 4 |
| ENSP00000335574 | zf-C2H2 | PF00096.26 | 7.4e-35 | 1 | 5 |
| ENSP00000335574 | zf-C2H2 | PF00096.26 | 7.4e-35 | 2 | 5 |
| ENSP00000335574 | zf-C2H2 | PF00096.26 | 7.4e-35 | 3 | 5 |
| ENSP00000335574 | zf-C2H2 | PF00096.26 | 7.4e-35 | 4 | 5 |
| ENSP00000335574 | zf-C2H2 | PF00096.26 | 7.4e-35 | 5 | 5 |
| ENSP00000419511 | zf-C2H2 | PF00096.26 | 1.4e-27 | 1 | 4 |
| ENSP00000419511 | zf-C2H2 | PF00096.26 | 1.4e-27 | 2 | 4 |
| ENSP00000419511 | zf-C2H2 | PF00096.26 | 1.4e-27 | 3 | 4 |
| ENSP00000419511 | zf-C2H2 | PF00096.26 | 1.4e-27 | 4 | 4 |
| ENSP00000483108 | zf-C2H2 | PF00096.26 | 3.4e-15 | 1 | 4 |
| ENSP00000483108 | zf-C2H2 | PF00096.26 | 3.4e-15 | 2 | 4 |
| ENSP00000483108 | zf-C2H2 | PF00096.26 | 3.4e-15 | 3 | 4 |
| ENSP00000483108 | zf-C2H2 | PF00096.26 | 3.4e-15 | 4 | 4 |
| ENSP00000420299 | zf-C2H2 | PF00096.26 | 3.5e-15 | 1 | 3 |
| ENSP00000420299 | zf-C2H2 | PF00096.26 | 3.5e-15 | 2 | 3 |
| ENSP00000420299 | zf-C2H2 | PF00096.26 | 3.5e-15 | 3 | 3 |
| ENSP00000420519 | zf-C2H2 | PF00096.26 | 1.9e-10 | 1 | 2 |
| ENSP00000420519 | zf-C2H2 | PF00096.26 | 1.9e-10 | 2 | 2 |
| ENSP00000305560 | zf-met | PF12874.7 | 8.9e-08 | 1 | 1 |
| ENSP00000369265 | zf-met | PF12874.7 | 8.9e-08 | 1 | 1 |
| ENSP00000420299 | zf-met | PF12874.7 | 2.9e-07 | 1 | 2 |
| ENSP00000420299 | zf-met | PF12874.7 | 2.9e-07 | 2 | 2 |
| ENSP00000420519 | zf-met | PF12874.7 | 3.7e-05 | 1 | 2 |
| ENSP00000420519 | zf-met | PF12874.7 | 3.7e-05 | 2 | 2 |
| ENSP00000369270 | zf-met | PF12874.7 | 0.00017 | 1 | 1 |
| ENSP00000483108 | zf-met | PF12874.7 | 0.0007 | 1 | 1 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 28341660 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000483150 | RREB1-208 | 2833 | - | ENSP00000419511 | 778 (aa) | - | C9JU34 |
| ENST00000379933 | RREB1-203 | 5826 | - | ENSP00000369265 | 1687 (aa) | - | Q92766 |
| ENST00000379938 | RREB1-204 | 8778 | - | ENSP00000369270 | 1742 (aa) | - | Q92766 |
| ENST00000471433 | RREB1-206 | 1021 | - | ENSP00000420299 | 164 (aa) | - | C9JPJ6 |
| ENST00000334984 | RREB1-201 | 5789 | - | ENSP00000335574 | 1476 (aa) | - | Q92766 |
| ENST00000491191 | RREB1-209 | 998 | - | ENSP00000420519 | 136 (aa) | - | C9JE09 |
| ENST00000611109 | RREB1-210 | 3563 | - | ENSP00000483108 | 755 (aa) | - | A0A087X055 |
| ENST00000475946 | RREB1-207 | 9067 | - | - | - (aa) | - | - |
| ENST00000349384 | RREB1-202 | 7440 | - | ENSP00000305560 | 1687 (aa) | - | Q92766 |
| ENST00000467782 | RREB1-205 | 243 | - | ENSP00000420571 | 4 (aa) | - | UPI0001B793E2 |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000124782 | Parkinson Disease | 4.356362E-8 | 17052657 |
| ENSG00000124782 | Social Support | 3.1540000E-005 | - |
| ENSG00000124782 | Platelet Function Tests | 4.5300000E-006 | - |
| ENSG00000124782 | Platelet Function Tests | 9.9400000E-006 | - |
| ENSG00000124782 | Adipose Tissue | 1E-8 | 27918534 |
| ENSG00000124782 | Body Mass Index | 1E-8 | 27918534 |
| ENSG00000124782 | Waist-Hip Ratio | 3E-8 | 23966867 |
| ENSG00000124782 | Multiple System Atrophy | 3E-6 | 27629089 |
| ENSG00000124782 | Glucose | 3E-7 | 22581228 |
| ENSG00000124782 | Body Mass Index | 3E-7 | 22581228 |
| ENSG00000124782 | Adipose Tissue | 6E-9 | 27918534 |
| ENSG00000124782 | Body Mass Index | 6E-9 | 27918534 |
| ENSG00000124782 | Diabetes Mellitus, Type 2 | 1E-9 | 24509480 |
| ENSG00000124782 | Optic Disk | 2E-8 | 28073927 |
| ENSG00000124782 | Macular Degeneration | 1E-6 | 20385826 |
| ENSG00000124782 | Adipose Tissue | 2E-6 | 27918534 |
| ENSG00000124782 | Body Mass Index | 2E-6 | 27918534 |
| ENSG00000124782 | Adipose Tissue | 4E-6 | 22589738 |
| ENSG00000124782 | Body Mass Index | 4E-6 | 22589738 |
| ENSG00000124782 | Adipose Tissue | 4E-6 | 22589738 |
| ENSG00000124782 | Body Mass Index | 4E-6 | 22589738 |
| ENSG00000124782 | Multiple Sclerosis | 3E-6 | 21833088 |
| ENSG00000124782 | Body Mass Index | 9E-6 | 27918534 |
| ENSG00000124782 | Subcutaneous Fat, Abdominal | 9E-6 | 27918534 |
| ENSG00000124782 | Optic Disk | 2E-10 | 28073927 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000124782 | rs557074 | 6 | 7144958 | G | Monocyte count | 27863252 | [0.022-0.04] unit increase | 0.03105644 | EFO_0005091 |
| ENSG00000124782 | rs55986493 | 6 | 7156526 | T | Monocyte count | 27863252 | [0.029-0.057] unit increase | 0.04291104 | EFO_0005091 |
| ENSG00000124782 | rs57996771 | 6 | 7157434 | C | Reticulocyte count | 27863252 | [0.023-0.043] unit increase | 0.03306844 | EFO_0007986 |
| ENSG00000124782 | rs75757892 | 6 | 7232156 | T | Red blood cell count | 27863252 | [0.023-0.041] unit increase | 0.03181995 | EFO_0004305 |
| ENSG00000124782 | rs11755724 | 6 | 7118757 | A | Multiple sclerosis | 21833088 | [1.06-1.09] | 1.08 | EFO_0003885 |
| ENSG00000124782 | rs1413700 | 6 | 7216251 | G | Multiple system atrophy | 27629089 | 1.39 | EFO_1001050 | |
| ENSG00000124782 | rs11755724 | 6 | 7118757 | G | Age-related macular degeneration | 20385826 | [1.09-1.14] | 1.15 | EFO_0001365 |
| ENSG00000124782 | rs6931262 | 6 | 7217284 | T | Waist-hip ratio | 23966867 | [0.04-0.08] unit increase | 0.06 | EFO_0004343 |
| ENSG00000124782 | rs2714337 | 6 | 7240344 | A | Fasting blood glucose | 22581228 | [0.0071-0.0173] unit decrease | 0.0122 | EFO_0004465 |
| ENSG00000124782 | rs2714337 | 6 | 7240344 | A | Fasting blood glucose (BMI interaction) | 22581228 | [-0.00087-0.00141] unit decrease | 2.68E-04 | EFO_0004340|EFO_0004465 |
| ENSG00000124782 | rs9379084 | 6 | 7231610 | G | Heel bone mineral density | 28869591 | [0.024-0.056] unit increase | 0.0397905 | EFO_0009270 |
| ENSG00000124782 | rs9379084 | 6 | 7231610 | G | Heel bone mineral density | 28869591 | [0.031-0.052] unit increase | 0.0413825 | EFO_0009270 |
| ENSG00000124782 | rs9379084 | 6 | 7231610 | G | Heel bone mineral density | 28869591 | [0.027-0.056] unit increase | 0.0410614 | EFO_0009270 |
| ENSG00000124782 | rs75757892 | 6 | 7232156 | T | Self-reported math ability (MTAG) | 30038396 | [0.0087-0.0181] unit increase | 0.0134 | EFO_0004875 |
| ENSG00000124782 | rs147287945 | 6 | 7223333 | G | Eotaxin levels | 27989323 | [0.081-0.204] SD units increase | 0.1428 | EFO_0008122 |
| ENSG00000124782 | rs75757892 | 6 | 7232156 | T | Hematocrit | 27863252 | [0.025-0.043] unit increase | 0.03442745 | EFO_0004348 |
| ENSG00000124782 | rs1285886 | 6 | 7140598 | A | Lymphocyte counts | 27863252 | [0.02-0.038] unit increase | 0.02893744 | EFO_0004587 |
| ENSG00000124782 | rs55986493 | 6 | 7156526 | T | Myeloid white cell count | 27863252 | [0.029-0.057] unit increase | 0.04286195 | EFO_0007988 |
| ENSG00000124782 | rs57996771 | 6 | 7157434 | C | Reticulocyte fraction of red cells | 27863252 | [0.023-0.043] unit increase | 0.03274342 | EFO_0007986 |
| ENSG00000124782 | rs11303054 | 6 | 7138928 | C | White blood cell count | 27863252 | [0.021-0.038] unit increase | 0.02959356 | EFO_0004308 |
| ENSG00000124782 | rs57996771 | 6 | 7157434 | C | High light scatter reticulocyte count | 27863252 | [0.02-0.039] unit increase | 0.02934395 | EFO_0007986 |
| ENSG00000124782 | rs4960295 | 6 | 7205563 | A | Vertical cup-disc ratio | 28073927 | [0.005-0.009] unit increase | 0.007 | EFO_0006939 |
| ENSG00000124782 | rs4960295 | 6 | 7205563 | A | Vertical cup-disc ratio | 28073927 | [0.005-0.009] unit increase | 0.007 | EFO_0006939 |
| ENSG00000124782 | rs1334577 | 6 | 7211518 | A | Neutrophil count | 27863252 | [0.017-0.033] unit decrease | 0.02485225 | EFO_0004833 |
| ENSG00000124782 | rs1334577 | 6 | 7211518 | A | Sum basophil neutrophil counts | 27863252 | [0.017-0.033] unit decrease | 0.02474309 | EFO_0004833|EFO_0005090 |
| ENSG00000124782 | rs9379084 | 6 | 7231610 | ? | Heel bone mineral density | 30048462 | [0.032-0.045] unit increase | 0.0389254 | EFO_0009270 |
| ENSG00000124782 | rs2842352 | 6 | 7250804 | ? | Heel bone mineral density | 30048462 | [0.013-0.025] unit increase | 0.0189983 | EFO_0009270 |
| ENSG00000124782 | rs9379084 | 6 | 7231610 | G | Heel bone mineral density | 30598549 | [0.028-0.04] unit increase | 0.0340199 | EFO_0009270 |
| ENSG00000124782 | rs3904600 | 6 | 7109432 | ? | Balding type 1 | 30595370 | EFO_0007825 | ||
| ENSG00000124782 | rs2714338 | 6 | 7250727 | ? | Balding type 1 | 30595370 | EFO_0007825 | ||
| ENSG00000124782 | rs9379084 | 6 | 7231610 | ? | Type 2 diabetes | 30595370 | EFO_0001360 | ||
| ENSG00000124782 | rs687467 | 6 | 7127843 | ? | Lung function (FVC) | 30595370 | EFO_0004312 | ||
| ENSG00000124782 | rs3904600 | 6 | 7109432 | ? | Waist-hip ratio | 30595370 | EFO_0004343 | ||
| ENSG00000124782 | rs1285886 | 6 | 7140598 | ? | White blood cell count | 30595370 | EFO_0004308 | ||
| ENSG00000124782 | rs75757892 | 6 | 7232156 | ? | Red blood cell count | 30595370 | EFO_0004305 | ||
| ENSG00000124782 | rs4960289 | 6 | 7146117 | ? | Lung function (FEV1/FVC) | 30595370 | EFO_0004713 | ||
| ENSG00000124782 | rs1334576 | 6 | 7211585 | A | Pulse pressure | 30224653 | [0.081-0.149] unit decrease | 0.1149 | EFO_0005763 |
| ENSG00000124782 | rs76052955 | 6 | 7213674 | A | Pulse pressure | 30578418 | [0.11-0.21] mmHg increase | 0.1613 | EFO_0005763 |
| ENSG00000124782 | rs9379084 | 6 | 7231610 | ? | Heel bone mineral density | 30595370 | EFO_0009270 | ||
| ENSG00000124782 | rs9379084 | 6 | 7231610 | A | Breast cancer | 29059683 | [0.028-0.069] unit increase | 0.0486 | EFO_0000305 |
| ENSG00000124782 | rs1334576 | 6 | 7211585 | G | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.013-0.021] unit increase | 0.0171 | EFO_0007788 |
| ENSG00000124782 | rs1334576 | 6 | 7211585 | G | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.012-0.021] unit increase | 0.0166 | EFO_0007788 |
| ENSG00000124782 | rs1334576 | 6 | 7211585 | G | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.015-0.027] unit increase | 0.0209 | EFO_0007788 |
| ENSG00000124782 | rs1334576 | 6 | 7211585 | G | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.0079-0.0201] unit increase | 0.014 | EFO_0007788 |
| ENSG00000124782 | rs1334576 | 6 | 7211585 | G | Waist-to-hip ratio adjusted for BMI (additive genetic model) | 30778226 | [0.014-0.026] unit increase | 0.0202 | EFO_0007788 |
| ENSG00000124782 | rs9379084 | 6 | 7231610 | G | Type 2 diabetes | 29632382 | [1.06-1.11] | 1.08 | EFO_0001360 |
| ENSG00000124782 | rs9379084 | 6 | 7231610 | G | Type 2 diabetes (adjusted for BMI) | 29632382 | [1.07-1.13] | 1.1 | EFO_0001360 |
| ENSG00000124782 | rs9379084 | 6 | 7231610 | G | Type 2 diabetes | 29632382 | [1.07-1.13] | 1.1 | EFO_0001360 |
| ENSG00000124782 | rs9379084 | 6 | 7231610 | G | Type 2 diabetes (adjusted for BMI) | 29632382 | [1.06-1.12] | 1.09 | EFO_0001360 |
| ENSG00000124782 | rs35742417 | 6 | 7247111 | C | Type 2 diabetes | 29632382 | [1.03-1.06] | 1.04 | EFO_0001360 |
| ENSG00000124782 | rs35742417 | 6 | 7247111 | C | Type 2 diabetes (adjusted for BMI) | 29632382 | [1.02-1.06] | 1.04 | EFO_0001360 |
| ENSG00000124782 | rs35742417 | 6 | 7247111 | C | Type 2 diabetes | 29632382 | (1.02-1.06) | 1.04 | EFO_0001360 |
| ENSG00000124782 | rs35742417 | 6 | 7247111 | C | Type 2 diabetes (adjusted for BMI) | 29632382 | [1.02-1.06] | 1.04 | EFO_0001360 |
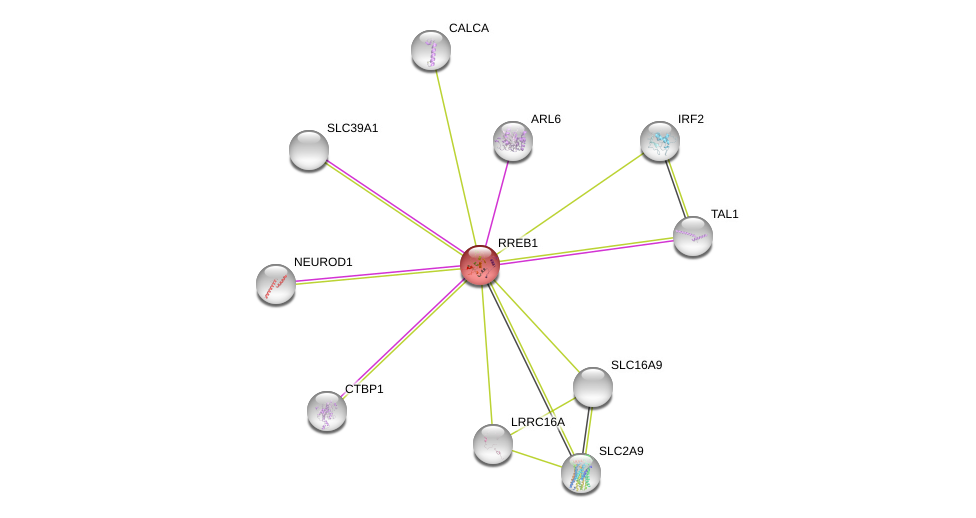
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000124782 | RREB1 | 59 | 42.982 | ENSAPEG00000002682 | - | 73 | 45.902 | Amphiprion_percula |
| ENSG00000124782 | RREB1 | 97 | 80.000 | ENSAPLG00000004726 | RREB1 | 100 | 63.754 | Anas_platyrhynchos |
| ENSG00000124782 | RREB1 | 97 | 74.016 | ENSACAG00000000520 | RREB1 | 100 | 61.051 | Anolis_carolinensis |
| ENSG00000124782 | RREB1 | 100 | 98.529 | ENSANAG00000029040 | RREB1 | 100 | 94.792 | Aotus_nancymaae |
| ENSG00000124782 | RREB1 | 99 | 67.470 | ENSAMXG00000019028 | rreb1a | 94 | 45.559 | Astyanax_mexicanus |
| ENSG00000124782 | RREB1 | 76 | 73.600 | ENSAMXG00000015314 | rreb1b | 96 | 43.486 | Astyanax_mexicanus |
| ENSG00000124782 | RREB1 | 100 | 98.780 | ENSCJAG00000021358 | RREB1 | 100 | 95.475 | Callithrix_jacchus |
| ENSG00000124782 | RREB1 | 100 | 96.341 | ENSCAFG00000009527 | RREB1 | 100 | 88.180 | Canis_familiaris |
| ENSG00000124782 | RREB1 | 100 | 96.341 | ENSCAFG00020013260 | RREB1 | 100 | 81.492 | Canis_lupus_dingo |
| ENSG00000124782 | RREB1 | 100 | 98.780 | ENSCCAG00000021905 | RREB1 | 100 | 95.249 | Cebus_capucinus |
| ENSG00000124782 | RREB1 | 100 | 99.390 | ENSCATG00000043662 | RREB1 | 100 | 98.263 | Cercocebus_atys |
| ENSG00000124782 | RREB1 | 100 | 99.390 | ENSCSAG00000007376 | RREB1 | 100 | 96.670 | Chlorocebus_sabaeus |
| ENSG00000124782 | RREB1 | 99 | 86.667 | ENSCPBG00000008709 | RREB1 | 100 | 65.133 | Chrysemys_picta_bellii |
| ENSG00000124782 | RREB1 | 100 | 99.390 | ENSCANG00000032835 | RREB1 | 100 | 96.979 | Colobus_angolensis_palliatus |
| ENSG00000124782 | RREB1 | 91 | 57.386 | ENSCSEG00000018342 | rreb1a | 88 | 50.980 | Cynoglossus_semilaevis |
| ENSG00000124782 | RREB1 | 95 | 63.910 | ENSDARG00000063701 | rreb1a | 92 | 62.245 | Danio_rerio |
| ENSG00000124782 | RREB1 | 100 | 96.341 | ENSDORG00000006979 | Rreb1 | 99 | 87.793 | Dipodomys_ordii |
| ENSG00000124782 | RREB1 | 100 | 96.341 | ENSEASG00005012054 | RREB1 | 100 | 82.379 | Equus_asinus_asinus |
| ENSG00000124782 | RREB1 | 100 | 96.341 | ENSECAG00000018169 | RREB1 | 94 | 88.713 | Equus_caballus |
| ENSG00000124782 | RREB1 | 98 | 86.466 | ENSFALG00000013231 | RREB1 | 96 | 63.673 | Ficedula_albicollis |
| ENSG00000124782 | RREB1 | 100 | 95.588 | ENSFDAG00000004213 | RREB1 | 96 | 68.487 | Fukomys_damarensis |
| ENSG00000124782 | RREB1 | 100 | 84.756 | ENSGALG00000012799 | RREB1 | 100 | 63.093 | Gallus_gallus |
| ENSG00000124782 | RREB1 | 92 | 77.080 | ENSGAGG00000002213 | RREB1 | 100 | 63.074 | Gopherus_agassizii |
| ENSG00000124782 | RREB1 | 100 | 99.486 | ENSGGOG00000004561 | RREB1 | 100 | 99.169 | Gorilla_gorilla |
| ENSG00000124782 | RREB1 | 93 | 62.092 | ENSLBEG00000013512 | rreb1a | 85 | 50.980 | Labrus_bergylta |
| ENSG00000124782 | RREB1 | 99 | 77.439 | ENSLACG00000009283 | RREB1 | 99 | 54.340 | Latimeria_chalumnae |
| ENSG00000124782 | RREB1 | 93 | 76.562 | ENSLOCG00000011702 | rreb1b | 96 | 51.724 | Lepisosteus_oculatus |
| ENSG00000124782 | RREB1 | 100 | 93.382 | ENSLAFG00000005300 | RREB1 | 100 | 78.100 | Loxodonta_africana |
| ENSG00000124782 | RREB1 | 100 | 99.390 | ENSMFAG00000000389 | RREB1 | 100 | 98.187 | Macaca_fascicularis |
| ENSG00000124782 | RREB1 | 100 | 99.390 | ENSMMUG00000007852 | RREB1 | 100 | 97.961 | Macaca_mulatta |
| ENSG00000124782 | RREB1 | 100 | 99.390 | ENSMNEG00000038556 | RREB1 | 100 | 98.187 | Macaca_nemestrina |
| ENSG00000124782 | RREB1 | 100 | 99.390 | ENSMLEG00000040965 | RREB1 | 100 | 98.187 | Mandrillus_leucophaeus |
| ENSG00000124782 | RREB1 | 88 | 88.966 | ENSMGAG00000003910 | RREB1 | 100 | 62.829 | Meleagris_gallopavo |
| ENSG00000124782 | RREB1 | 100 | 95.244 | ENSMICG00000003683 | RREB1 | 100 | 91.053 | Microcebus_murinus |
| ENSG00000124782 | RREB1 | 100 | 89.706 | ENSMODG00000009892 | RREB1 | 100 | 70.922 | Monodelphis_domestica |
| ENSG00000124782 | RREB1 | 100 | 95.732 | ENSMPUG00000000558 | RREB1 | 100 | 82.452 | Mustela_putorius_furo |
| ENSG00000124782 | RREB1 | 100 | 96.341 | ENSMLUG00000013153 | RREB1 | 100 | 81.935 | Myotis_lucifugus |
| ENSG00000124782 | RREB1 | 100 | 96.341 | ENSNGAG00000021506 | Rreb1 | 100 | 83.737 | Nannospalax_galili |
| ENSG00000124782 | RREB1 | 74 | 72.143 | ENSNBRG00000016744 | rreb1a | 85 | 54.082 | Neolamprologus_brichardi |
| ENSG00000124782 | RREB1 | 100 | 99.390 | ENSNLEG00000011554 | RREB1 | 100 | 99.018 | Nomascus_leucogenys |
| ENSG00000124782 | RREB1 | 100 | 88.415 | ENSMEUG00000015377 | RREB1 | 100 | 72.629 | Notamacropus_eugenii |
| ENSG00000124782 | RREB1 | 100 | 90.441 | ENSOPRG00000001966 | RREB1 | 88 | 77.527 | Ochotona_princeps |
| ENSG00000124782 | RREB1 | 100 | 85.403 | ENSODEG00000007124 | RREB1 | 100 | 77.876 | Octodon_degus |
| ENSG00000124782 | RREB1 | 88 | 61.194 | ENSOMEG00000001389 | rreb1a | 68 | 94.118 | Oryzias_melastigma |
| ENSG00000124782 | RREB1 | 100 | 95.732 | ENSOGAG00000025755 | RREB1 | 100 | 81.353 | Otolemur_garnettii |
| ENSG00000124782 | RREB1 | 100 | 99.743 | ENSPPAG00000037017 | RREB1 | 100 | 99.698 | Pan_paniscus |
| ENSG00000124782 | RREB1 | 100 | 96.341 | ENSPPRG00000012144 | RREB1 | 100 | 83.771 | Panthera_pardus |
| ENSG00000124782 | RREB1 | 100 | 99.743 | ENSPTRG00000017697 | RREB1 | 100 | 99.622 | Pan_troglodytes |
| ENSG00000124782 | RREB1 | 100 | 99.390 | ENSPANG00000019553 | RREB1 | 100 | 97.961 | Papio_anubis |
| ENSG00000124782 | RREB1 | 92 | 63.934 | ENSPKIG00000006372 | rreb1b | 91 | 53.403 | Paramormyrops_kingsleyae |
| ENSG00000124782 | RREB1 | 88 | 90.345 | ENSPSIG00000006582 | RREB1 | 100 | 63.435 | Pelodiscus_sinensis |
| ENSG00000124782 | RREB1 | 100 | 94.118 | ENSPEMG00000014153 | Rreb1 | 100 | 80.201 | Peromyscus_maniculatus_bairdii |
| ENSG00000124782 | RREB1 | 92 | 82.418 | ENSPCIG00000012644 | RREB1 | 100 | 70.413 | Phascolarctos_cinereus |
| ENSG00000124782 | RREB1 | 100 | 99.100 | ENSPPYG00000016204 | RREB1 | 100 | 97.819 | Pongo_abelii |
| ENSG00000124782 | RREB1 | 100 | 96.324 | ENSPCOG00000014821 | RREB1 | 100 | 91.573 | Propithecus_coquereli |
| ENSG00000124782 | RREB1 | 100 | 96.341 | ENSPVAG00000014081 | RREB1 | 100 | 82.924 | Pteropus_vampyrus |
| ENSG00000124782 | RREB1 | 96 | 65.409 | ENSPNAG00000000486 | rreb1a | 89 | 51.600 | Pygocentrus_nattereri |
| ENSG00000124782 | RREB1 | 100 | 93.382 | ENSRNOG00000015701 | Rreb1 | 100 | 79.263 | Rattus_norvegicus |
| ENSG00000124782 | RREB1 | 100 | 99.390 | ENSRROG00000039603 | RREB1 | 100 | 97.508 | Rhinopithecus_roxellana |
| ENSG00000124782 | RREB1 | 100 | 98.780 | ENSSBOG00000025257 | RREB1 | 100 | 94.800 | Saimiri_boliviensis_boliviensis |
| ENSG00000124782 | RREB1 | 100 | 88.971 | ENSSHAG00000005295 | RREB1 | 91 | 83.184 | Sarcophilus_harrisii |
| ENSG00000124782 | RREB1 | 88 | 70.470 | ENSSFOG00015001117 | rreb1b | 93 | 48.257 | Scleropages_formosus |
| ENSG00000124782 | RREB1 | 88 | 88.276 | ENSSPUG00000007686 | RREB1 | 100 | 63.300 | Sphenodon_punctatus |
| ENSG00000124782 | RREB1 | 100 | 84.756 | ENSTGUG00000002309 | RREB1 | 100 | 65.530 | Taeniopygia_guttata |
| ENSG00000124782 | RREB1 | 100 | 95.122 | ENSUAMG00000003741 | RREB1 | 100 | 82.864 | Ursus_americanus |
| ENSG00000124782 | RREB1 | 100 | 96.341 | ENSVVUG00000005050 | RREB1 | 96 | 88.444 | Vulpes_vulpes |
| ENSG00000124782 | RREB1 | 96 | 81.529 | ENSXETG00000015435 | rreb1 | 98 | 55.556 | Xenopus_tropicalis |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000122 | negative regulation of transcription by RNA polymerase II | 21873635. | IBA | Process |
| GO:0000122 | negative regulation of transcription by RNA polymerase II | 20133935. | IMP | Process |
| GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | 21873635. | IBA | Function |
| GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | 8816445. | IDA | Function |
| GO:0000978 | RNA polymerase II proximal promoter sequence-specific DNA binding | 21873635. | IBA | Function |
| GO:0000978 | RNA polymerase II proximal promoter sequence-specific DNA binding | 20133935. | IMP | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | - | ISA | Function |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 19274049. | NAS | Function |
| GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | 21873635. | IBA | Function |
| GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | 8816445. | IDA | Function |
| GO:0001650 | fibrillar center | - | IDA | Component |
| GO:0003700 | DNA-binding transcription factor activity | 21873635. | IBA | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 9305772. | TAS | Component |
| GO:0005737 | cytoplasm | 9305772. | TAS | Component |
| GO:0006355 | regulation of transcription, DNA-templated | 9305772. | TAS | Process |
| GO:0006366 | transcription by RNA polymerase II | 8816445. | TAS | Process |
| GO:0007265 | Ras protein signal transduction | 8816445. | TAS | Process |
| GO:0007275 | multicellular organism development | 8816445. | TAS | Process |
| GO:0010634 | positive regulation of epithelial cell migration | 18394891. | IMP | Process |
| GO:0016604 | nuclear body | 9305772. | TAS | Component |
| GO:0016607 | nuclear speck | - | IDA | Component |
| GO:0033601 | positive regulation of mammary gland epithelial cell proliferation | 18394891. | IMP | Process |
| GO:0045893 | positive regulation of transcription, DNA-templated | 9305772. | TAS | Process |
| GO:0045944 | positive regulation of transcription by RNA polymerase II | 8816445. | IDA | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0070062 | extracellular exosome | 19056867. | HDA | Component |
| GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 18394891. | IMP | Process |
| GO:1903691 | positive regulation of wound healing, spreading of epidermal cells | 18394891. | IMP | Process |
| GO:2000394 | positive regulation of lamellipodium morphogenesis | 18394891. | IMP | Process |