
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| PRAD | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| CHOL | |||
| SKCM | |||
| TGCT | |||
| UCEC |
| Cancer | P-value | Q-value |
|---|---|---|
| SARC | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| SKCM | ||
| BRCA | ||
| ESCA | ||
| KIRP | ||
| COAD | ||
| LAML | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| LUAD | ||
| OV |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 29458450 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000494627 | GOT2-204 | 889 | - | - | - (aa) | - | - |
| ENST00000496461 | GOT2-205 | 981 | - | - | - (aa) | - | - |
| ENST00000492378 | GOT2-203 | 987 | - | - | - (aa) | - | - |
| ENST00000564400 | GOT2-206 | 983 | - | - | - (aa) | - | - |
| ENST00000434819 | GOT2-202 | 1615 | - | ENSP00000394100 | 387 (aa) | - | P00505 |
| ENST00000245206 | GOT2-201 | 2462 | - | ENSP00000245206 | 430 (aa) | - | P00505 |
| ENST00000568368 | GOT2-207 | 516 | - | ENSP00000456205 | 59 (aa) | - | H3BRE7 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa00220 | Arginine biosynthesis | KEGG |
| hsa00250 | Alanine, aspartate and glutamate metabolism | KEGG |
| hsa00270 | Cysteine and methionine metabolism | KEGG |
| hsa00330 | Arginine and proline metabolism | KEGG |
| hsa00350 | Tyrosine metabolism | KEGG |
| hsa00360 | Phenylalanine metabolism | KEGG |
| hsa00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | KEGG |
| hsa01100 | Metabolic pathways | KEGG |
| hsa01200 | Carbon metabolism | KEGG |
| hsa01210 | 2-Oxocarboxylic acid metabolism | KEGG |
| hsa01230 | Biosynthesis of amino acids | KEGG |
| hsa04975 | Fat digestion and absorption | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000125166 | rs73550818 | 16 | 58730951 | ? | Aspartate aminotransferase levels | 29403010 | [0.021-0.037] unit increase novel | 0.02855 | EFO_0004736 |
| ENSG00000125166 | rs12600277 | 16 | 58725428 | ? | Aortic root size | 21223598 | [0.082-0.198] cm increase | 0.1399 | EFO_0005037 |
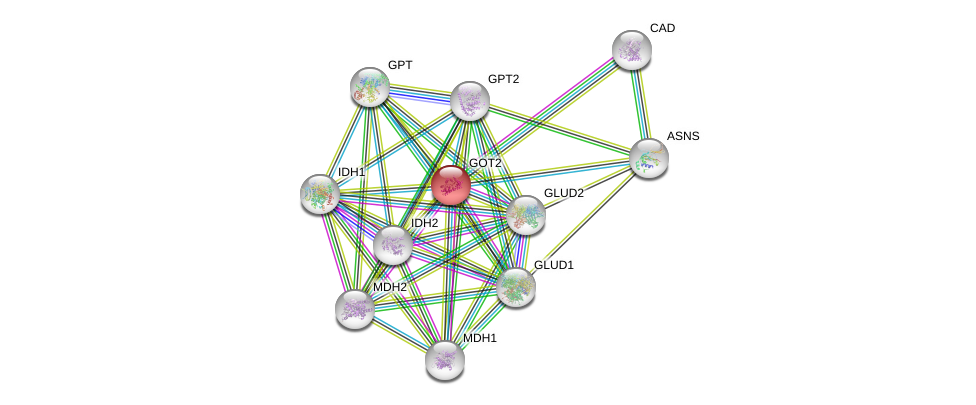
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000125166 | GOT2 | 93 | 43.284 | WBGene00020146 | got-1.2 | 97 | 43.577 | Caenorhabditis_elegans |
| ENSG00000125166 | GOT2 | 96 | 63.107 | FBgn0001125 | Got2 | 96 | 63.107 | Drosophila_melanogaster |
| ENSG00000125166 | GOT2 | 95 | 48.058 | FBgn0001124 | Got1 | 95 | 48.866 | Drosophila_melanogaster |
| ENSG00000125166 | GOT2 | 93 | 48.894 | ENSG00000120053 | GOT1 | 98 | 48.889 | Homo_sapiens |
| ENSG00000125166 | GOT2 | 100 | 94.651 | ENSMUSG00000031672 | Got2 | 100 | 94.651 | Mus_musculus |
| ENSG00000125166 | GOT2 | 93 | 43.659 | YLR027C | AAT2 | 97 | 44.089 | Saccharomyces_cerevisiae |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity | 21873635. | IBA | Function |
| GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity | 2182221.2567216.2731362. | IDA | Function |
| GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity | - | ISS | Function |
| GO:0005543 | phospholipid binding | - | IEA | Function |
| GO:0005739 | mitochondrion | 20833797. | HDA | Component |
| GO:0005739 | mitochondrion | 21873635. | IBA | Component |
| GO:0005739 | mitochondrion | 9537447. | IDA | Component |
| GO:0005739 | mitochondrion | - | ISS | Component |
| GO:0005743 | mitochondrial inner membrane | - | IEA | Component |
| GO:0005759 | mitochondrial matrix | - | TAS | Component |
| GO:0005886 | plasma membrane | 9537447. | IDA | Component |
| GO:0006094 | gluconeogenesis | - | TAS | Process |
| GO:0006103 | 2-oxoglutarate metabolic process | - | ISS | Process |
| GO:0006107 | oxaloacetate metabolic process | - | IEA | Process |
| GO:0006531 | aspartate metabolic process | - | ISS | Process |
| GO:0006532 | aspartate biosynthetic process | - | IEA | Process |
| GO:0006533 | aspartate catabolic process | 21873635. | IBA | Process |
| GO:0006533 | aspartate catabolic process | 2567216. | IDA | Process |
| GO:0006536 | glutamate metabolic process | - | ISS | Process |
| GO:0007565 | female pregnancy | - | IEA | Process |
| GO:0007595 | lactation | - | IEA | Process |
| GO:0008652 | cellular amino acid biosynthetic process | - | TAS | Process |
| GO:0009986 | cell surface | - | IEA | Component |
| GO:0014850 | response to muscle activity | - | IEA | Process |
| GO:0015908 | fatty acid transport | 9537447. | IEP | Process |
| GO:0016212 | kynurenine-oxoglutarate transaminase activity | - | IEA | Function |
| GO:0016597 | amino acid binding | - | IEA | Function |
| GO:0019470 | 4-hydroxyproline catabolic process | 21998747. | TAS | Process |
| GO:0019550 | glutamate catabolic process to aspartate | - | IEA | Process |
| GO:0019551 | glutamate catabolic process to 2-oxoglutarate | - | IEA | Process |
| GO:0019899 | enzyme binding | - | IEA | Function |
| GO:0030170 | pyridoxal phosphate binding | - | IEA | Function |
| GO:0030315 | T-tubule | - | IEA | Component |
| GO:0032868 | response to insulin | - | IEA | Process |
| GO:0032991 | protein-containing complex | - | IEA | Component |
| GO:0042803 | protein homodimerization activity | - | IEA | Function |
| GO:0043204 | perikaryon | - | IEA | Component |
| GO:0043278 | response to morphine | - | IEA | Process |
| GO:0045471 | response to ethanol | 9537447. | IDA | Process |
| GO:0046487 | glyoxylate metabolic process | - | TAS | Process |
| GO:0070062 | extracellular exosome | 20458337.23533145. | HDA | Component |
| GO:0097052 | L-kynurenine metabolic process | - | IEA | Process |