
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 25519054 |
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| CHOL | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| THCA | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UVM |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| ACC | |||
| BLCA | |||
| BRCA | |||
| CESC | |||
| HNSC | |||
| KIRC | |||
| KIRC | |||
| KIRP | |||
| LGG | |||
| LIHC | |||
| LUSC | |||
| OV | |||
| PAAD | |||
| PAAD | |||
| SARC | |||
| SKCM | |||
| TGCT | |||
| THCA | |||
| UCEC | |||
| UVM |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| MESO | ||
| SKCM | ||
| BRCA | ||
| CESC | ||
| GBM | ||
| LGG | ||
| LUAD | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000346169 | MMR_HSR1 | PF01926.23 | 0.00019 | 1 | 1 |
| ENSP00000353278 | MMR_HSR1 | PF01926.23 | 0.00019 | 1 | 1 |
| ENSP00000378108 | MMR_HSR1 | PF01926.23 | 0.00019 | 1 | 1 |
| ENSP00000418715 | MMR_HSR1 | PF01926.23 | 0.00019 | 1 | 1 |
| ENSP00000341524 | MMR_HSR1 | PF01926.23 | 0.00019 | 1 | 1 |
| ENSP00000346397 | MMR_HSR1 | PF01926.23 | 0.00019 | 1 | 1 |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 17229681 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000467310 | SEPT6-207 | 1061 | - | - | - (aa) | - | - |
| ENST00000354416 | SEPT6-203 | 4652 | XM_005262400 | ENSP00000346397 | 431 (aa) | XP_005262457 | B1AMS2 |
| ENST00000481072 | SEPT6-208 | 836 | - | - | - (aa) | - | - |
| ENST00000360156 | SEPT6-204 | 2609 | XM_006724750 | ENSP00000353278 | 427 (aa) | XP_006724813 | Q14141 |
| ENST00000489216 | SEPT6-209 | 1617 | - | ENSP00000418715 | 427 (aa) | - | Q14141 |
| ENST00000343984 | SEPT6-201 | 2693 | XM_011531318 | ENSP00000341524 | 434 (aa) | XP_011529620 | Q14141 |
| ENST00000394610 | SEPT6-205 | 4694 | - | ENSP00000378108 | 427 (aa) | - | Q14141 |
| ENST00000460411 | SEPT6-206 | 1393 | - | ENSP00000435818 | 267 (aa) | - | Q14141 |
| ENST00000354228 | SEPT6-202 | 1576 | - | ENSP00000346169 | 429 (aa) | - | Q14141 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa05100 | Bacterial invasion of epithelial cells | KEGG |
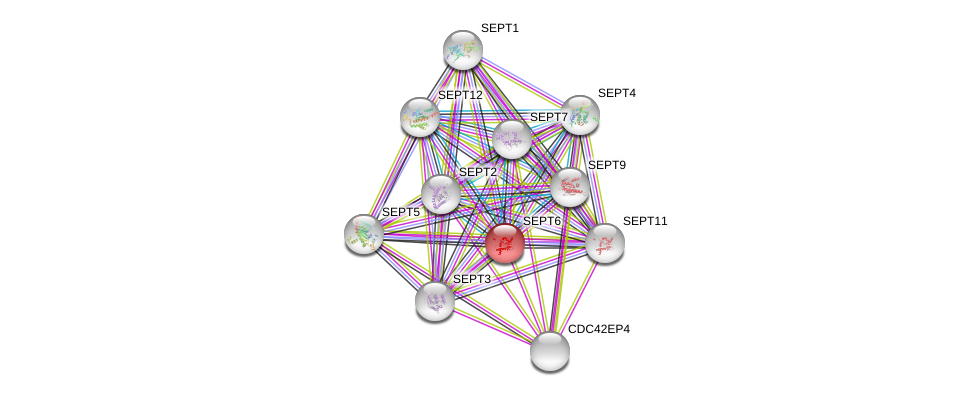
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000125354 | SEPT6 | 100 | 90.398 | ENSAPOG00000000347 | sept6 | 100 | 90.398 | Acanthochromis_polyacanthus |
| ENSG00000125354 | SEPT6 | 99 | 90.141 | ENSACIG00000013682 | sept6 | 99 | 90.141 | Amphilophus_citrinellus |
| ENSG00000125354 | SEPT6 | 100 | 90.398 | ENSAOCG00000011293 | sept6 | 100 | 90.398 | Amphiprion_ocellaris |
| ENSG00000125354 | SEPT6 | 100 | 90.398 | ENSAPEG00000005460 | sept6 | 100 | 90.398 | Amphiprion_percula |
| ENSG00000125354 | SEPT6 | 100 | 89.930 | ENSATEG00000018814 | sept6 | 100 | 89.930 | Anabas_testudineus |
| ENSG00000125354 | SEPT6 | 100 | 99.539 | ENSANAG00000033580 | SEPT6 | 100 | 99.539 | Aotus_nancymaae |
| ENSG00000125354 | SEPT6 | 100 | 90.164 | ENSACLG00000001734 | sept6 | 100 | 90.164 | Astatotilapia_calliptera |
| ENSG00000125354 | SEPT6 | 100 | 90.210 | ENSAMXG00000033660 | sept6 | 100 | 90.210 | Astyanax_mexicanus |
| ENSG00000125354 | SEPT6 | 100 | 99.539 | ENSCJAG00000005022 | SEPT6 | 100 | 99.539 | Callithrix_jacchus |
| ENSG00000125354 | SEPT6 | 100 | 91.935 | ENSCCAG00000023266 | SEPT6 | 100 | 91.935 | Cebus_capucinus |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSCATG00000036060 | SEPT6 | 100 | 100.000 | Cercocebus_atys |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSCSAG00000018351 | SEPT6 | 100 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSCANG00000016430 | SEPT6 | 100 | 100.000 | Colobus_angolensis_palliatus |
| ENSG00000125354 | SEPT6 | 100 | 98.376 | ENSCGRG00001014207 | Sept6 | 100 | 98.376 | Cricetulus_griseus_chok1gshd |
| ENSG00000125354 | SEPT6 | 99 | 97.387 | ENSCGRG00000007825 | Sept6 | 100 | 98.113 | Cricetulus_griseus_crigri |
| ENSG00000125354 | SEPT6 | 100 | 90.398 | ENSCSEG00000002026 | sept6 | 94 | 90.398 | Cynoglossus_semilaevis |
| ENSG00000125354 | SEPT6 | 100 | 90.398 | ENSCVAG00000004959 | sept6 | 100 | 90.398 | Cyprinodon_variegatus |
| ENSG00000125354 | SEPT6 | 100 | 90.632 | ENSDARG00000010721 | sept6 | 99 | 90.632 | Danio_rerio |
| ENSG00000125354 | SEPT6 | 100 | 90.867 | ENSELUG00000020499 | sept6 | 96 | 90.867 | Esox_lucius |
| ENSG00000125354 | SEPT6 | 100 | 98.157 | ENSFCAG00000008744 | SEPT6 | 100 | 98.157 | Felis_catus |
| ENSG00000125354 | SEPT6 | 100 | 98.157 | ENSFDAG00000002473 | SEPT6 | 100 | 98.157 | Fukomys_damarensis |
| ENSG00000125354 | SEPT6 | 100 | 90.398 | ENSFHEG00000017987 | sept6 | 100 | 90.398 | Fundulus_heteroclitus |
| ENSG00000125354 | SEPT6 | 100 | 89.862 | ENSGMOG00000001671 | sept6 | 100 | 89.862 | Gadus_morhua |
| ENSG00000125354 | SEPT6 | 100 | 90.164 | ENSGAFG00000000922 | sept6 | 100 | 90.164 | Gambusia_affinis |
| ENSG00000125354 | SEPT6 | 100 | 90.092 | ENSGACG00000017441 | sept6 | 100 | 90.092 | Gasterosteus_aculeatus |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSGGOG00000009733 | SEPT6 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000125354 | SEPT6 | 100 | 90.164 | ENSHBUG00000017506 | sept6 | 100 | 90.164 | Haplochromis_burtoni |
| ENSG00000125354 | SEPT6 | 100 | 90.443 | ENSHCOG00000017047 | sept6 | 100 | 90.443 | Hippocampus_comes |
| ENSG00000125354 | SEPT6 | 100 | 90.443 | ENSIPUG00000019317 | sept6 | 85 | 90.443 | Ictalurus_punctatus |
| ENSG00000125354 | SEPT6 | 100 | 97.696 | ENSJJAG00000020412 | Sept6 | 100 | 97.696 | Jaculus_jaculus |
| ENSG00000125354 | SEPT6 | 100 | 90.210 | ENSKMAG00000020207 | sept6 | 100 | 90.210 | Kryptolebias_marmoratus |
| ENSG00000125354 | SEPT6 | 100 | 90.443 | ENSLBEG00000028683 | sept6 | 95 | 90.443 | Labrus_bergylta |
| ENSG00000125354 | SEPT6 | 100 | 91.101 | ENSLOCG00000015233 | sept6 | 100 | 91.771 | Lepisosteus_oculatus |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSMFAG00000003084 | SEPT6 | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSMMUG00000017669 | SEPT6 | 100 | 100.000 | Macaca_mulatta |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSMNEG00000026932 | SEPT6 | 100 | 100.000 | Macaca_nemestrina |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSMLEG00000030428 | SEPT6 | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000125354 | SEPT6 | 100 | 90.164 | ENSMAMG00000019988 | sept6 | 100 | 90.164 | Mastacembelus_armatus |
| ENSG00000125354 | SEPT6 | 100 | 90.164 | ENSMZEG00005007935 | sept6 | 89 | 90.164 | Maylandia_zebra |
| ENSG00000125354 | SEPT6 | 100 | 98.157 | ENSMAUG00000018038 | Sept6 | 100 | 98.157 | Mesocricetus_auratus |
| ENSG00000125354 | SEPT6 | 98 | 98.571 | ENSMOCG00000000500 | Sept6 | 99 | 98.798 | Microtus_ochrogaster |
| ENSG00000125354 | SEPT6 | 97 | 91.063 | ENSMMOG00000004176 | sept6 | 97 | 91.063 | Mola_mola |
| ENSG00000125354 | SEPT6 | 100 | 89.631 | ENSMALG00000007921 | sept6 | 100 | 89.631 | Monopterus_albus |
| ENSG00000125354 | SEPT6 | 100 | 96.503 | MGP_CAROLIEiJ_G0032996 | Sept6 | 100 | 96.503 | Mus_caroli |
| ENSG00000125354 | SEPT6 | 100 | 96.270 | ENSMUSG00000050379 | Sept6 | 100 | 96.270 | Mus_musculus |
| ENSG00000125354 | SEPT6 | 99 | 94.737 | MGP_PahariEiJ_G0031540 | - | 71 | 95.817 | Mus_pahari |
| ENSG00000125354 | SEPT6 | 100 | 96.503 | MGP_SPRETEiJ_G0034149 | Sept6 | 100 | 96.503 | Mus_spretus |
| ENSG00000125354 | SEPT6 | 97 | 97.115 | ENSMLUG00000005623 | SEPT6 | 95 | 97.115 | Myotis_lucifugus |
| ENSG00000125354 | SEPT6 | 100 | 96.544 | ENSNGAG00000010702 | Sept6 | 100 | 96.544 | Nannospalax_galili |
| ENSG00000125354 | SEPT6 | 99 | 82.864 | ENSNBRG00000011675 | sept6 | 99 | 82.864 | Neolamprologus_brichardi |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSNLEG00000014873 | SEPT6 | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000125354 | SEPT6 | 100 | 90.398 | ENSONIG00000002528 | sept6 | 99 | 90.398 | Oreochromis_niloticus |
| ENSG00000125354 | SEPT6 | 100 | 89.930 | ENSORLG00000000997 | sept6 | 100 | 90.750 | Oryzias_latipes |
| ENSG00000125354 | SEPT6 | 100 | 89.930 | ENSORLG00020003886 | sept6 | 100 | 89.930 | Oryzias_latipes_hni |
| ENSG00000125354 | SEPT6 | 100 | 89.930 | ENSORLG00015013025 | sept6 | 100 | 89.930 | Oryzias_latipes_hsok |
| ENSG00000125354 | SEPT6 | 100 | 88.759 | ENSOMEG00000014650 | sept6 | 100 | 88.759 | Oryzias_melastigma |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSPPAG00000040865 | SEPT6 | 100 | 100.000 | Pan_paniscus |
| ENSG00000125354 | SEPT6 | 100 | 97.926 | ENSPPRG00000023588 | SEPT6 | 100 | 97.926 | Panthera_pardus |
| ENSG00000125354 | SEPT6 | 93 | 97.177 | ENSPTIG00000015338 | SEPT6 | 99 | 84.428 | Panthera_tigris_altaica |
| ENSG00000125354 | SEPT6 | 100 | 99.770 | ENSPTRG00000022227 | SEPT6 | 100 | 99.770 | Pan_troglodytes |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSPANG00000000899 | SEPT6 | 100 | 100.000 | Papio_anubis |
| ENSG00000125354 | SEPT6 | 100 | 85.315 | ENSPKIG00000019114 | sept6 | 100 | 85.315 | Paramormyrops_kingsleyae |
| ENSG00000125354 | SEPT6 | 99 | 91.549 | ENSPKIG00000013948 | SEPT6 | 100 | 92.250 | Paramormyrops_kingsleyae |
| ENSG00000125354 | SEPT6 | 99 | 88.498 | ENSPMGG00000012133 | sept6 | 100 | 88.498 | Periophthalmus_magnuspinnatus |
| ENSG00000125354 | SEPT6 | 100 | 98.387 | ENSPEMG00000016450 | Sept6 | 100 | 98.387 | Peromyscus_maniculatus_bairdii |
| ENSG00000125354 | SEPT6 | 100 | 90.783 | ENSPCIG00000028910 | SEPT6 | 100 | 90.783 | Phascolarctos_cinereus |
| ENSG00000125354 | SEPT6 | 98 | 90.736 | ENSPFOG00000024721 | sept6 | 97 | 90.736 | Poecilia_formosa |
| ENSG00000125354 | SEPT6 | 84 | 88.000 | ENSPLAG00000005147 | sept6 | 93 | 88.000 | Poecilia_latipinna |
| ENSG00000125354 | SEPT6 | 100 | 90.398 | ENSPLAG00000019878 | sept6 | 100 | 90.398 | Poecilia_latipinna |
| ENSG00000125354 | SEPT6 | 100 | 90.398 | ENSPMEG00000023470 | sept6 | 100 | 90.398 | Poecilia_mexicana |
| ENSG00000125354 | SEPT6 | 100 | 89.930 | ENSPREG00000012310 | sept6 | 100 | 89.930 | Poecilia_reticulata |
| ENSG00000125354 | SEPT6 | 100 | 98.126 | ENSPPYG00000020673 | SEPT6 | 100 | 98.126 | Pongo_abelii |
| ENSG00000125354 | SEPT6 | 100 | 90.210 | ENSPNYG00000009709 | sept6 | 100 | 91.000 | Pundamilia_nyererei |
| ENSG00000125354 | SEPT6 | 100 | 90.909 | ENSPNAG00000021638 | sept6 | 100 | 90.909 | Pygocentrus_nattereri |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSRBIG00000044672 | SEPT6 | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000125354 | SEPT6 | 100 | 100.000 | ENSRROG00000038667 | SEPT6 | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000125354 | SEPT6 | 100 | 99.309 | ENSSBOG00000024143 | SEPT6 | 100 | 99.309 | Saimiri_boliviensis_boliviensis |
| ENSG00000125354 | SEPT6 | 99 | 90.588 | ENSSHAG00000007460 | SEPT6 | 98 | 90.588 | Sarcophilus_harrisii |
| ENSG00000125354 | SEPT6 | 100 | 90.443 | ENSSFOG00015010832 | SEPT6 | 100 | 90.443 | Scleropages_formosus |
| ENSG00000125354 | SEPT6 | 100 | 88.759 | ENSSFOG00015005477 | sept6 | 100 | 88.759 | Scleropages_formosus |
| ENSG00000125354 | SEPT6 | 100 | 90.092 | ENSSMAG00000012785 | sept6 | 100 | 90.092 | Scophthalmus_maximus |
| ENSG00000125354 | SEPT6 | 100 | 90.398 | ENSSDUG00000005575 | sept6 | 100 | 90.398 | Seriola_dumerili |
| ENSG00000125354 | SEPT6 | 100 | 90.443 | ENSSLDG00000003277 | sept6 | 100 | 90.443 | Seriola_lalandi_dorsalis |
| ENSG00000125354 | SEPT6 | 100 | 90.398 | ENSSPAG00000010108 | sept6 | 100 | 90.398 | Stegastes_partitus |
| ENSG00000125354 | SEPT6 | 100 | 89.696 | ENSTRUG00000010830 | sept6 | 100 | 90.796 | Takifugu_rubripes |
| ENSG00000125354 | SEPT6 | 100 | 88.290 | ENSTNIG00000015761 | sept6 | 100 | 88.167 | Tetraodon_nigroviridis |
| ENSG00000125354 | SEPT6 | 90 | 84.583 | ENSXCOG00000016712 | sept6 | 89 | 84.583 | Xiphophorus_couchianus |
| ENSG00000125354 | SEPT6 | 100 | 90.443 | ENSXMAG00000013699 | sept6 | 100 | 90.443 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000281 | mitotic cytokinesis | 11809673. | TAS | Process |
| GO:0000777 | condensed chromosome kinetochore | - | IEA | Component |
| GO:0003924 | GTPase activity | 21873635. | IBA | Function |
| GO:0005515 | protein binding | 16189514.17229681.17637674.17803907.18047794.18330356.24633326.25416956.25588830.25775403.26871637.30021884. | IPI | Function |
| GO:0005525 | GTP binding | 11809673. | TAS | Function |
| GO:0005819 | spindle | - | IEA | Component |
| GO:0005940 | septin ring | 21873635. | IBA | Component |
| GO:0007283 | spermatogenesis | - | IEA | Process |
| GO:0008021 | synaptic vesicle | - | IEA | Component |
| GO:0015630 | microtubule cytoskeleton | 21873635. | IBA | Component |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0030154 | cell differentiation | - | IEA | Process |
| GO:0030496 | midbody | - | IEA | Component |
| GO:0031105 | septin complex | 21873635. | IBA | Component |
| GO:0031105 | septin complex | 25588830. | IDA | Component |
| GO:0032154 | cleavage furrow | - | IEA | Component |
| GO:0032173 | septin collar | - | IEA | Component |
| GO:0043679 | axon terminus | - | IEA | Component |
| GO:0060090 | molecular adaptor activity | 21873635. | IBA | Function |
| GO:0060271 | cilium assembly | 21873635. | IBA | Process |
| GO:0061640 | cytoskeleton-dependent cytokinesis | 21873635. | IBA | Process |
| GO:0097227 | sperm annulus | 25588830. | IDA | Component |