
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| PAAD | |||
| READ | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| PRAD | ||
| LUSC | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| BLCA | ||
| UCEC | ||
| LIHC | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000252542 | RRM_1 | PF00076.22 | 1.7e-13 | 1 | 1 |
| ENSP00000252542 | SAP | PF02037.27 | 6.4e-10 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000252542 | SAFB2-201 | 3371 | XM_011528449 | ENSP00000252542 | 953 (aa) | XP_011526751 | Q14151 |
| ENST00000585449 | SAFB2-202 | 443 | - | - | - (aa) | - | - |
| ENST00000589925 | SAFB2-204 | 506 | - | - | - (aa) | - | - |
| ENST00000590262 | SAFB2-206 | 564 | - | ENSP00000468454 | 124 (aa) | - | K7ERX3 |
| ENST00000591101 | SAFB2-207 | 551 | - | - | - (aa) | - | - |
| ENST00000591310 | SAFB2-210 | 722 | - | - | - (aa) | - | - |
| ENST00000590000 | SAFB2-205 | 633 | - | - | - (aa) | - | - |
| ENST00000587802 | SAFB2-203 | 583 | - | - | - (aa) | - | - |
| ENST00000592599 | SAFB2-211 | 2260 | - | - | - (aa) | - | - |
| ENST00000591120 | SAFB2-208 | 1298 | - | ENSP00000468505 | 115 (aa) | - | Q14151 |
| ENST00000591123 | SAFB2-209 | 836 | - | ENSP00000467981 | 87 (aa) | - | K7EQU4 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000130254 | rs113123495 | 19 | 5587529 | C | Obsessive-compulsive symptoms | 26859814 | unit increase | 10.582 | EFO_0007802 |
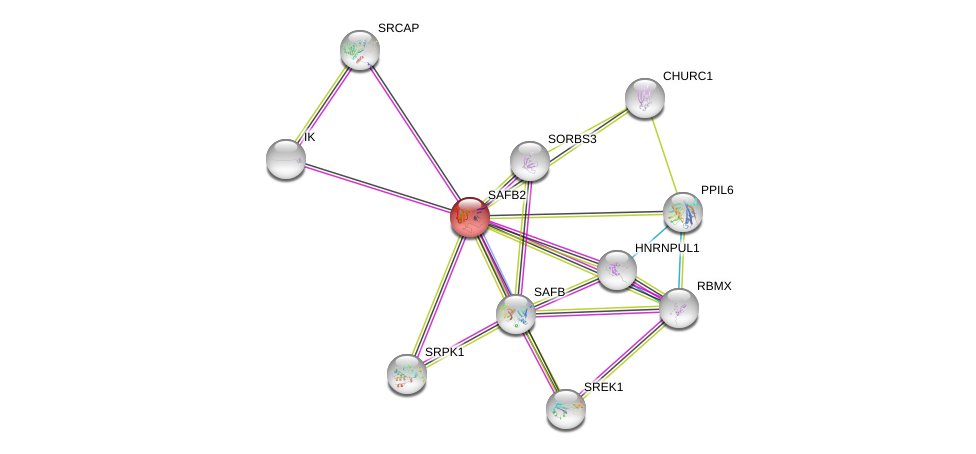
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000130254 | SAFB2 | 57 | 50.769 | ENSG00000137776 | SLTM | 88 | 47.561 |
| ENSG00000130254 | SAFB2 | 98 | 69.313 | ENSG00000160633 | SAFB | 100 | 69.874 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000130254 | SAFB2 | 100 | 86.254 | ENSAMEG00000012146 | SAFB2 | 100 | 86.149 | Ailuropoda_melanoleuca |
| ENSG00000130254 | SAFB2 | 100 | 96.642 | ENSANAG00000031936 | SAFB2 | 100 | 96.642 | Aotus_nancymaae |
| ENSG00000130254 | SAFB2 | 100 | 84.067 | ENSBTAG00000001790 | SAFB2 | 99 | 84.509 | Bos_taurus |
| ENSG00000130254 | SAFB2 | 100 | 92.742 | ENSCJAG00000017205 | - | 97 | 92.093 | Callithrix_jacchus |
| ENSG00000130254 | SAFB2 | 100 | 88.796 | ENSCAFG00000018814 | SAFB2 | 100 | 88.796 | Canis_familiaris |
| ENSG00000130254 | SAFB2 | 100 | 88.796 | ENSCAFG00020018939 | SAFB2 | 100 | 88.796 | Canis_lupus_dingo |
| ENSG00000130254 | SAFB2 | 100 | 83.648 | ENSCHIG00000024547 | SAFB2 | 100 | 83.648 | Capra_hircus |
| ENSG00000130254 | SAFB2 | 93 | 67.245 | ENSCAPG00000011771 | SAFB2 | 100 | 67.795 | Cavia_aperea |
| ENSG00000130254 | SAFB2 | 100 | 79.124 | ENSCPOG00000011394 | SAFB2 | 100 | 79.226 | Cavia_porcellus |
| ENSG00000130254 | SAFB2 | 100 | 96.747 | ENSCCAG00000017348 | SAFB2 | 100 | 96.747 | Cebus_capucinus |
| ENSG00000130254 | SAFB2 | 100 | 97.272 | ENSCATG00000039497 | SAFB2 | 100 | 97.272 | Cercocebus_atys |
| ENSG00000130254 | SAFB2 | 100 | 98.321 | ENSCSAG00000009959 | SAFB2 | 100 | 98.321 | Chlorocebus_sabaeus |
| ENSG00000130254 | SAFB2 | 87 | 77.778 | ENSCHOG00000010684 | SAFB2 | 99 | 76.412 | Choloepus_hoffmanni |
| ENSG00000130254 | SAFB2 | 100 | 95.908 | ENSCANG00000024675 | SAFB2 | 100 | 95.908 | Colobus_angolensis_palliatus |
| ENSG00000130254 | SAFB2 | 100 | 70.080 | ENSCGRG00001015833 | Safb2 | 100 | 69.779 | Cricetulus_griseus_chok1gshd |
| ENSG00000130254 | SAFB2 | 100 | 70.080 | ENSCGRG00000011131 | Safb2 | 100 | 69.779 | Cricetulus_griseus_crigri |
| ENSG00000130254 | SAFB2 | 88 | 74.312 | ENSDNOG00000045791 | - | 99 | 81.321 | Dasypus_novemcinctus |
| ENSG00000130254 | SAFB2 | 95 | 75.840 | ENSDORG00000016199 | Safb2 | 100 | 76.056 | Dipodomys_ordii |
| ENSG00000130254 | SAFB2 | 80 | 79.348 | ENSETEG00000016756 | - | 50 | 85.650 | Echinops_telfairi |
| ENSG00000130254 | SAFB2 | 86 | 83.636 | ENSEASG00005022952 | SAFB2 | 91 | 83.135 | Equus_asinus_asinus |
| ENSG00000130254 | SAFB2 | 98 | 83.565 | ENSECAG00000018978 | SAFB2 | 100 | 83.721 | Equus_caballus |
| ENSG00000130254 | SAFB2 | 100 | 87.631 | ENSFCAG00000011834 | SAFB2 | 100 | 87.631 | Felis_catus |
| ENSG00000130254 | SAFB2 | 93 | 82.780 | ENSFDAG00000020009 | SAFB2 | 93 | 83.065 | Fukomys_damarensis |
| ENSG00000130254 | SAFB2 | 56 | 62.500 | ENSGAGG00000008422 | - | 86 | 62.500 | Gopherus_agassizii |
| ENSG00000130254 | SAFB2 | 100 | 96.455 | ENSGGOG00000015093 | SAFB2 | 100 | 96.455 | Gorilla_gorilla |
| ENSG00000130254 | SAFB2 | 100 | 85.905 | ENSHGLG00000017442 | SAFB2 | 100 | 86.550 | Heterocephalus_glaber_female |
| ENSG00000130254 | SAFB2 | 98 | 82.632 | ENSLAFG00000003836 | SAFB2 | 99 | 81.770 | Loxodonta_africana |
| ENSG00000130254 | SAFB2 | 100 | 98.531 | ENSMFAG00000039706 | SAFB2 | 100 | 98.531 | Macaca_fascicularis |
| ENSG00000130254 | SAFB2 | 100 | 98.636 | ENSMMUG00000017968 | SAFB2 | 100 | 98.636 | Macaca_mulatta |
| ENSG00000130254 | SAFB2 | 100 | 92.476 | ENSMNEG00000032042 | SAFB2 | 100 | 92.092 | Macaca_nemestrina |
| ENSG00000130254 | SAFB2 | 100 | 98.741 | ENSMLEG00000034562 | SAFB2 | 100 | 98.741 | Mandrillus_leucophaeus |
| ENSG00000130254 | SAFB2 | 82 | 75.784 | ENSMAUG00000011573 | Safb2 | 92 | 76.302 | Mesocricetus_auratus |
| ENSG00000130254 | SAFB2 | 100 | 91.004 | ENSMICG00000008711 | SAFB2 | 100 | 91.004 | Microcebus_murinus |
| ENSG00000130254 | SAFB2 | 100 | 69.122 | ENSMOCG00000006827 | Safb2 | 100 | 68.681 | Microtus_ochrogaster |
| ENSG00000130254 | SAFB2 | 98 | 72.334 | ENSMODG00000001178 | SAFB2 | 100 | 71.813 | Monodelphis_domestica |
| ENSG00000130254 | SAFB2 | 100 | 69.384 | MGP_CAROLIEiJ_G0021779 | Safb2 | 100 | 69.085 | Mus_caroli |
| ENSG00000130254 | SAFB2 | 100 | 69.552 | ENSMUSG00000042625 | Safb2 | 100 | 69.353 | Mus_musculus |
| ENSG00000130254 | SAFB2 | 100 | 69.000 | MGP_PahariEiJ_G0020768 | Safb2 | 100 | 68.700 | Mus_pahari |
| ENSG00000130254 | SAFB2 | 100 | 64.121 | MGP_SPRETEiJ_G0022690 | Safb2 | 100 | 62.157 | Mus_spretus |
| ENSG00000130254 | SAFB2 | 100 | 87.120 | ENSMPUG00000006106 | SAFB2 | 93 | 87.016 | Mustela_putorius_furo |
| ENSG00000130254 | SAFB2 | 100 | 82.884 | ENSMLUG00000001050 | SAFB2 | 100 | 81.224 | Myotis_lucifugus |
| ENSG00000130254 | SAFB2 | 100 | 97.692 | ENSNLEG00000013514 | SAFB2 | 100 | 97.692 | Nomascus_leucogenys |
| ENSG00000130254 | SAFB2 | 93 | 68.568 | ENSMEUG00000002456 | SAFB2 | 100 | 68.233 | Notamacropus_eugenii |
| ENSG00000130254 | SAFB2 | 100 | 75.495 | ENSOPRG00000011523 | - | 100 | 75.600 | Ochotona_princeps |
| ENSG00000130254 | SAFB2 | 100 | 82.280 | ENSODEG00000014751 | SAFB2 | 100 | 82.176 | Octodon_degus |
| ENSG00000130254 | SAFB2 | 97 | 49.587 | ENSOANG00000011319 | - | 97 | 53.881 | Ornithorhynchus_anatinus |
| ENSG00000130254 | SAFB2 | 98 | 50.820 | ENSOANG00000013644 | - | 100 | 53.531 | Ornithorhynchus_anatinus |
| ENSG00000130254 | SAFB2 | 97 | 60.833 | ENSOCUG00000001592 | - | 98 | 77.622 | Oryctolagus_cuniculus |
| ENSG00000130254 | SAFB2 | 100 | 89.749 | ENSOGAG00000006855 | SAFB2 | 100 | 88.935 | Otolemur_garnettii |
| ENSG00000130254 | SAFB2 | 97 | 74.689 | ENSOARG00000007982 | SAFB2 | 67 | 84.976 | Ovis_aries |
| ENSG00000130254 | SAFB2 | 100 | 99.790 | ENSPPAG00000043392 | SAFB2 | 100 | 99.790 | Pan_paniscus |
| ENSG00000130254 | SAFB2 | 100 | 87.526 | ENSPPRG00000011310 | SAFB2 | 100 | 87.526 | Panthera_pardus |
| ENSG00000130254 | SAFB2 | 82 | 87.862 | ENSPTIG00000015120 | SAFB2 | 91 | 87.862 | Panthera_tigris_altaica |
| ENSG00000130254 | SAFB2 | 100 | 100.000 | ENSPTRG00000010334 | SAFB2 | 100 | 93.704 | Pan_troglodytes |
| ENSG00000130254 | SAFB2 | 100 | 98.636 | ENSPANG00000007004 | SAFB2 | 100 | 98.636 | Papio_anubis |
| ENSG00000130254 | SAFB2 | 100 | 67.697 | ENSPEMG00000016015 | Safb2 | 100 | 67.398 | Peromyscus_maniculatus_bairdii |
| ENSG00000130254 | SAFB2 | 64 | 75.000 | ENSPCIG00000009518 | - | 99 | 78.637 | Phascolarctos_cinereus |
| ENSG00000130254 | SAFB2 | 100 | 99.056 | ENSPPYG00000009427 | SAFB2 | 100 | 99.056 | Pongo_abelii |
| ENSG00000130254 | SAFB2 | 100 | 80.104 | ENSPCAG00000006352 | SAFB2 | 99 | 81.654 | Procavia_capensis |
| ENSG00000130254 | SAFB2 | 100 | 87.631 | ENSPCOG00000023260 | SAFB2 | 100 | 85.879 | Propithecus_coquereli |
| ENSG00000130254 | SAFB2 | 98 | 74.380 | ENSPVAG00000004902 | - | 71 | 86.957 | Pteropus_vampyrus |
| ENSG00000130254 | SAFB2 | 82 | 69.417 | ENSRNOG00000049511 | AABR07066522.1 | 100 | 67.476 | Rattus_norvegicus |
| ENSG00000130254 | SAFB2 | 94 | 94.872 | ENSRBIG00000033446 | SAFB2 | 100 | 99.124 | Rhinopithecus_bieti |
| ENSG00000130254 | SAFB2 | 100 | 98.426 | ENSRROG00000006606 | SAFB2 | 100 | 98.426 | Rhinopithecus_roxellana |
| ENSG00000130254 | SAFB2 | 100 | 95.278 | ENSSBOG00000032315 | SAFB2 | 100 | 96.231 | Saimiri_boliviensis_boliviensis |
| ENSG00000130254 | SAFB2 | 98 | 71.932 | ENSSHAG00000012703 | SAFB2 | 100 | 70.668 | Sarcophilus_harrisii |
| ENSG00000130254 | SAFB2 | 81 | 72.165 | ENSSARG00000000649 | SAFB2 | 100 | 72.165 | Sorex_araneus |
| ENSG00000130254 | SAFB2 | 100 | 88.901 | ENSSSCG00000013523 | SAFB2 | 100 | 88.901 | Sus_scrofa |
| ENSG00000130254 | SAFB2 | 89 | 75.455 | ENSTBEG00000012709 | SAFB2 | 86 | 78.937 | Tupaia_belangeri |
| ENSG00000130254 | SAFB2 | 93 | 80.493 | ENSTTRG00000013585 | SAFB2 | 93 | 80.269 | Tursiops_truncatus |
| ENSG00000130254 | SAFB2 | 98 | 88.260 | ENSUAMG00000024695 | SAFB2 | 97 | 88.260 | Ursus_americanus |
| ENSG00000130254 | SAFB2 | 86 | 86.144 | ENSUMAG00000009585 | SAFB2 | 90 | 86.144 | Ursus_maritimus |
| ENSG00000130254 | SAFB2 | 89 | 78.182 | ENSVPAG00000012041 | SAFB2 | 89 | 90.625 | Vicugna_pacos |
| ENSG00000130254 | SAFB2 | 100 | 88.691 | ENSVVUG00000013695 | SAFB2 | 100 | 88.691 | Vulpes_vulpes |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22658674.22681889. | HDA | Function |
| GO:0005515 | protein binding | 12660241.20562859. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005737 | cytoplasm | - | IEA | Component |
| GO:0006357 | regulation of transcription by RNA polymerase II | 21873635. | IBA | Process |
| GO:0016604 | nuclear body | - | IDA | Component |
| GO:0042802 | identical protein binding | 19674106. | IPI | Function |
| GO:0043231 | intracellular membrane-bounded organelle | - | IDA | Component |
| GO:0043565 | sequence-specific DNA binding | 21873635. | IBA | Function |
| GO:0050684 | regulation of mRNA processing | 21873635. | IBA | Process |
| GO:0060008 | Sertoli cell differentiation | - | IEA | Process |
| GO:0060765 | regulation of androgen receptor signaling pathway | 21873635. | IBA | Process |
| GO:0070062 | extracellular exosome | 18570454. | HDA | Component |