
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| LIHC | |||
| PAAD | |||
| READ | |||
| THCA | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| STAD | ||
| MESO | ||
| ACC | ||
| PRAD | ||
| LUSC | ||
| PAAD | ||
| BLCA | ||
| CESC | ||
| UCEC | ||
| LIHC | ||
| UVM | ||
| OV |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000252622 | LSM | PF01423.22 | 1.4e-20 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000591515 | LSM7-206 | 710 | - | - | - (aa) | - | - |
| ENST00000591745 | LSM7-207 | 543 | - | - | - (aa) | - | - |
| ENST00000589532 | LSM7-205 | 539 | - | - | - (aa) | - | - |
| ENST00000252622 | LSM7-201 | 529 | XM_017026869 | ENSP00000252622 | 103 (aa) | XP_016882358 | Q9UK45 |
| ENST00000587502 | LSM7-204 | 575 | - | ENSP00000485007 | 49 (aa) | - | A0A087X2I5 |
| ENST00000585395 | LSM7-202 | 621 | - | ENSP00000466556 | 60 (aa) | - | K7EML7 |
| ENST00000585409 | LSM7-203 | 512 | - | ENSP00000467145 | 35 (aa) | - | K7ENY5 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000130332 | rs8103741 | 19 | 2325006 | A | Educational attainment (MTAG) | 30038396 | [0.0078-0.0144] unit decrease | 0.0111 | EFO_0004784 |
| ENSG00000130332 | rs8103741 | 19 | 2325006 | A | Educational attainment (years of education) | 30038396 | [0.0071-0.0145] unit decrease | 0.0108 | EFO_0004784 |
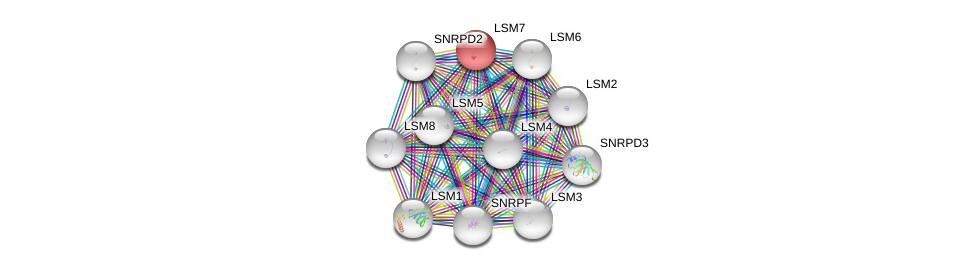
| Ensembl ID | Gene Symbol | Coverage | Identiy | Paralog | Gene Symbol | Coverage | Identiy |
|---|---|---|---|---|---|---|---|
| ENSG00000130332 | LSM7 | 72 | 35.526 | ENSG00000128534 | LSM8 | 73 | 35.526 |
| ENSG00000130332 | LSM7 | 69 | 36.620 | ENSG00000143977 | SNRPG | 91 | 39.394 |
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000130332 | LSM7 | 91 | 95.745 | ENSAPOG00000015573 | lsm7 | 81 | 95.745 | Acanthochromis_polyacanthus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSAMEG00000005484 | LSM7 | 100 | 97.087 | Ailuropoda_melanoleuca |
| ENSG00000130332 | LSM7 | 95 | 94.898 | ENSACIG00000006498 | lsm7 | 78 | 94.898 | Amphilophus_citrinellus |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSAOCG00000001077 | lsm7 | 92 | 95.050 | Amphiprion_ocellaris |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSAPEG00000024447 | lsm7 | 92 | 95.050 | Amphiprion_percula |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSAPEG00000000694 | lsm7 | 100 | 95.146 | Amphiprion_percula |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSATEG00000008557 | lsm7 | 100 | 95.146 | Anabas_testudineus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSANAG00000037196 | - | 100 | 100.000 | Aotus_nancymaae |
| ENSG00000130332 | LSM7 | 96 | 97.872 | ENSANAG00000022222 | - | 100 | 91.262 | Aotus_nancymaae |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSACLG00000021021 | lsm7 | 100 | 94.175 | Astatotilapia_calliptera |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSAMXG00000037622 | lsm7 | 100 | 95.146 | Astyanax_mexicanus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSBTAG00000004521 | LSM7 | 100 | 100.000 | Bos_taurus |
| ENSG00000130332 | LSM7 | 97 | 57.000 | WBGene00003081 | lsm-7 | 96 | 57.000 | Caenorhabditis_elegans |
| ENSG00000130332 | LSM7 | 94 | 96.970 | ENSCJAG00000039713 | - | 68 | 100.000 | Callithrix_jacchus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSCAFG00000019409 | LSM7 | 98 | 99.010 | Canis_familiaris |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSCAFG00020018168 | LSM7 | 74 | 99.010 | Canis_lupus_dingo |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSCHIG00000023663 | LSM7 | 100 | 98.058 | Capra_hircus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSCAPG00000014534 | LSM7 | 100 | 100.000 | Cavia_aperea |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSCPOG00000014025 | LSM7 | 100 | 100.000 | Cavia_porcellus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSCCAG00000035456 | LSM7 | 99 | 100.000 | Cebus_capucinus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSCATG00000023543 | LSM7 | 100 | 100.000 | Cercocebus_atys |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSCLAG00000011635 | LSM7 | 100 | 100.000 | Chinchilla_lanigera |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSCSAG00000011599 | LSM7 | 100 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSCPBG00000021317 | LSM7 | 77 | 94.444 | Chrysemys_picta_bellii |
| ENSG00000130332 | LSM7 | 94 | 80.435 | ENSCING00000022134 | - | 97 | 80.208 | Ciona_intestinalis |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSCANG00000035525 | LSM7 | 100 | 100.000 | Colobus_angolensis_palliatus |
| ENSG00000130332 | LSM7 | 100 | 99.029 | ENSCGRG00001000807 | - | 100 | 99.029 | Cricetulus_griseus_chok1gshd |
| ENSG00000130332 | LSM7 | 86 | 90.000 | ENSCGRG00001016005 | - | 92 | 88.889 | Cricetulus_griseus_chok1gshd |
| ENSG00000130332 | LSM7 | 100 | 97.959 | ENSCGRG00000014355 | - | 97 | 99.010 | Cricetulus_griseus_crigri |
| ENSG00000130332 | LSM7 | 86 | 90.000 | ENSCGRG00000006821 | - | 92 | 88.889 | Cricetulus_griseus_crigri |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSCSEG00000014900 | lsm7 | 86 | 95.050 | Cynoglossus_semilaevis |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSCVAG00000021091 | lsm7 | 94 | 96.040 | Cyprinodon_variegatus |
| ENSG00000130332 | LSM7 | 100 | 97.959 | ENSDARG00000058328 | lsm7 | 100 | 95.146 | Danio_rerio |
| ENSG00000130332 | LSM7 | 100 | 97.959 | ENSDORG00000024460 | Lsm7 | 100 | 98.039 | Dipodomys_ordii |
| ENSG00000130332 | LSM7 | 94 | 76.531 | FBgn0261068 | LSm7 | 89 | 76.531 | Drosophila_melanogaster |
| ENSG00000130332 | LSM7 | 100 | 97.959 | ENSETEG00000012788 | LSM7 | 99 | 97.030 | Echinops_telfairi |
| ENSG00000130332 | LSM7 | 100 | 84.466 | ENSEBUG00000016992 | lsm7 | 99 | 84.466 | Eptatretus_burgeri |
| ENSG00000130332 | LSM7 | 80 | 61.538 | ENSEBUG00000010470 | - | 79 | 59.036 | Eptatretus_burgeri |
| ENSG00000130332 | LSM7 | 100 | 96.117 | ENSECAG00000001440 | LSM7 | 66 | 96.117 | Equus_caballus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSFCAG00000003445 | LSM7 | 100 | 99.029 | Felis_catus |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSFALG00000011912 | LSM7 | 100 | 97.030 | Ficedula_albicollis |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSFDAG00000007661 | LSM7 | 100 | 100.000 | Fukomys_damarensis |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSFHEG00000006349 | lsm7 | 90 | 96.040 | Fundulus_heteroclitus |
| ENSG00000130332 | LSM7 | 100 | 91.837 | ENSGMOG00000013320 | lsm7 | 100 | 91.262 | Gadus_morhua |
| ENSG00000130332 | LSM7 | 100 | 96.117 | ENSGAFG00000013902 | lsm7 | 100 | 96.117 | Gambusia_affinis |
| ENSG00000130332 | LSM7 | 100 | 93.878 | ENSGACG00000010390 | lsm7 | 100 | 92.233 | Gasterosteus_aculeatus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSGGOG00000010172 | LSM7 | 100 | 100.000 | Gorilla_gorilla |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSHBUG00000004616 | lsm7 | 100 | 94.175 | Haplochromis_burtoni |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSHGLG00000010587 | LSM7 | 100 | 100.000 | Heterocephalus_glaber_female |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSHGLG00100000852 | LSM7 | 100 | 100.000 | Heterocephalus_glaber_male |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSHCOG00000020252 | lsm7 | 100 | 94.059 | Hippocampus_comes |
| ENSG00000130332 | LSM7 | 100 | 95.146 | ENSIPUG00000024724 | lsm7 | 100 | 95.146 | Ictalurus_punctatus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSSTOG00000028372 | LSM7 | 99 | 100.000 | Ictidomys_tridecemlineatus |
| ENSG00000130332 | LSM7 | 98 | 98.020 | ENSJJAG00000001936 | Lsm7 | 98 | 98.020 | Jaculus_jaculus |
| ENSG00000130332 | LSM7 | 100 | 96.117 | ENSKMAG00000022151 | lsm7 | 100 | 96.117 | Kryptolebias_marmoratus |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSLBEG00000005203 | lsm7 | 100 | 94.175 | Labrus_bergylta |
| ENSG00000130332 | LSM7 | 100 | 97.959 | ENSLACG00000015700 | LSM7 | 100 | 97.087 | Latimeria_chalumnae |
| ENSG00000130332 | LSM7 | 100 | 97.959 | ENSLOCG00000004356 | lsm7 | 100 | 96.117 | Lepisosteus_oculatus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSLAFG00000029302 | LSM7 | 98 | 100.000 | Loxodonta_africana |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSMFAG00000044572 | LSM7 | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSMMUG00000000452 | LSM7 | 100 | 100.000 | Macaca_mulatta |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSMNEG00000004393 | LSM7 | 100 | 100.000 | Macaca_nemestrina |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSMLEG00000030852 | LSM7 | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSMAMG00000005410 | lsm7 | 100 | 94.175 | Mastacembelus_armatus |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSMZEG00005028055 | lsm7 | 100 | 94.175 | Maylandia_zebra |
| ENSG00000130332 | LSM7 | 100 | 98.058 | ENSMGAG00000000750 | LSM7 | 89 | 98.058 | Meleagris_gallopavo |
| ENSG00000130332 | LSM7 | 100 | 99.029 | ENSMAUG00000009276 | Lsm7 | 100 | 99.029 | Mesocricetus_auratus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSMICG00000028619 | - | 100 | 100.000 | Microcebus_murinus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSMICG00000009988 | - | 100 | 99.029 | Microcebus_murinus |
| ENSG00000130332 | LSM7 | 100 | 99.029 | ENSMOCG00000016925 | Lsm7 | 100 | 99.029 | Microtus_ochrogaster |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSMMOG00000019645 | lsm7 | 98 | 95.146 | Mola_mola |
| ENSG00000130332 | LSM7 | 100 | 99.029 | ENSMODG00000004722 | LSM7 | 100 | 99.029 | Monodelphis_domestica |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSMALG00000014511 | lsm7 | 79 | 94.175 | Monopterus_albus |
| ENSG00000130332 | LSM7 | 100 | 97.087 | MGP_CAROLIEiJ_G0015607 | Lsm7 | 100 | 97.087 | Mus_caroli |
| ENSG00000130332 | LSM7 | 100 | 97.087 | ENSMUSG00000035215 | Lsm7 | 100 | 97.087 | Mus_musculus |
| ENSG00000130332 | LSM7 | 100 | 97.087 | MGP_PahariEiJ_G0031020 | Lsm7 | 100 | 97.087 | Mus_pahari |
| ENSG00000130332 | LSM7 | 100 | 97.087 | MGP_SPRETEiJ_G0016425 | Lsm7 | 100 | 97.087 | Mus_spretus |
| ENSG00000130332 | LSM7 | 100 | 92.233 | ENSMLUG00000024002 | - | 94 | 92.233 | Myotis_lucifugus |
| ENSG00000130332 | LSM7 | 100 | 97.959 | ENSMLUG00000013097 | - | 100 | 97.087 | Myotis_lucifugus |
| ENSG00000130332 | LSM7 | 100 | 98.058 | ENSNGAG00000021637 | Lsm7 | 100 | 98.058 | Nannospalax_galili |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSNBRG00000015770 | lsm7 | 87 | 94.059 | Neolamprologus_brichardi |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSNBRG00000001237 | - | 82 | 92.233 | Neolamprologus_brichardi |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSNLEG00000002265 | LSM7 | 100 | 100.000 | Nomascus_leucogenys |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSODEG00000013590 | - | 100 | 100.000 | Octodon_degus |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSODEG00000018769 | - | 100 | 93.204 | Octodon_degus |
| ENSG00000130332 | LSM7 | 100 | 87.755 | ENSODEG00000008190 | - | 100 | 79.612 | Octodon_degus |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSONIG00000001801 | lsm7 | 89 | 94.175 | Oreochromis_niloticus |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSORLG00000023951 | lsm7 | 100 | 95.146 | Oryzias_latipes |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSORLG00020022226 | lsm7 | 100 | 95.146 | Oryzias_latipes_hni |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSORLG00015020044 | lsm7 | 94 | 95.050 | Oryzias_latipes_hsok |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSOMEG00000012806 | lsm7 | 100 | 95.146 | Oryzias_melastigma |
| ENSG00000130332 | LSM7 | 100 | 88.350 | ENSOGAG00000024373 | - | 100 | 88.350 | Otolemur_garnettii |
| ENSG00000130332 | LSM7 | 98 | 98.020 | ENSOGAG00000000144 | - | 99 | 98.020 | Otolemur_garnettii |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSOARG00000014013 | LSM7 | 100 | 98.058 | Ovis_aries |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSPPAG00000026500 | LSM7 | 100 | 100.000 | Pan_paniscus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSPPRG00000008624 | LSM7 | 100 | 99.029 | Panthera_pardus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSPTIG00000016252 | LSM7 | 98 | 99.010 | Panthera_tigris_altaica |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSPTRG00000010233 | LSM7 | 100 | 100.000 | Pan_troglodytes |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSPANG00000020668 | LSM7 | 100 | 100.000 | Papio_anubis |
| ENSG00000130332 | LSM7 | 100 | 97.959 | ENSPKIG00000011325 | lsm7 | 100 | 97.087 | Paramormyrops_kingsleyae |
| ENSG00000130332 | LSM7 | 100 | 97.959 | ENSPSIG00000017149 | LSM7 | 89 | 98.020 | Pelodiscus_sinensis |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSPMGG00000006498 | lsm7 | 100 | 95.146 | Periophthalmus_magnuspinnatus |
| ENSG00000130332 | LSM7 | 100 | 99.029 | ENSPCIG00000030454 | LSM7 | 100 | 99.029 | Phascolarctos_cinereus |
| ENSG00000130332 | LSM7 | 100 | 93.878 | ENSPFOG00000012467 | lsm7 | 91 | 93.204 | Poecilia_formosa |
| ENSG00000130332 | LSM7 | 100 | 93.878 | ENSPLAG00000000252 | lsm7 | 94 | 94.118 | Poecilia_latipinna |
| ENSG00000130332 | LSM7 | 100 | 93.878 | ENSPMEG00000000587 | lsm7 | 94 | 94.118 | Poecilia_mexicana |
| ENSG00000130332 | LSM7 | 94 | 93.939 | ENSPREG00000016718 | lsm7 | 65 | 76.543 | Poecilia_reticulata |
| ENSG00000130332 | LSM7 | 98 | 100.000 | ENSPPYG00000009348 | - | 50 | 97.260 | Pongo_abelii |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSPCOG00000017177 | LSM7 | 100 | 100.000 | Propithecus_coquereli |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSPNYG00000006938 | lsm7 | 100 | 94.175 | Pundamilia_nyererei |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSPNAG00000010141 | lsm7 | 100 | 95.146 | Pygocentrus_nattereri |
| ENSG00000130332 | LSM7 | 100 | 98.058 | ENSRNOG00000019552 | Lsm7 | 100 | 98.058 | Rattus_norvegicus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSRBIG00000035978 | LSM7 | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSRROG00000034044 | LSM7 | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000130332 | LSM7 | 86 | 47.826 | YNL147W | - | 80 | 47.826 | Saccharomyces_cerevisiae |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSSBOG00000034899 | LSM7 | 100 | 100.000 | Saimiri_boliviensis_boliviensis |
| ENSG00000130332 | LSM7 | 100 | 95.146 | ENSSFOG00015006728 | lsm7 | 100 | 95.146 | Scleropages_formosus |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSSMAG00000004195 | lsm7 | 93 | 95.050 | Scophthalmus_maximus |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSSDUG00000001863 | lsm7 | 100 | 95.146 | Seriola_dumerili |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSSLDG00000016339 | lsm7 | 100 | 95.146 | Seriola_lalandi_dorsalis |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSSPAG00000015221 | lsm7 | 100 | 95.146 | Stegastes_partitus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSSSCG00000013463 | LSM7 | 99 | 100.000 | Sus_scrofa |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSTGUG00000000067 | LSM7 | 100 | 97.030 | Taeniopygia_guttata |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSTRUG00000022913 | lsm7 | 100 | 95.146 | Takifugu_rubripes |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSTNIG00000015525 | lsm7 | 100 | 95.146 | Tetraodon_nigroviridis |
| ENSG00000130332 | LSM7 | 100 | 85.714 | ENSTBEG00000016602 | LSM7 | 100 | 87.143 | Tupaia_belangeri |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSUAMG00000000912 | LSM7 | 86 | 97.087 | Ursus_americanus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSUMAG00000006556 | LSM7 | 79 | 99.010 | Ursus_maritimus |
| ENSG00000130332 | LSM7 | 100 | 100.000 | ENSVVUG00000002640 | LSM7 | 65 | 98.592 | Vulpes_vulpes |
| ENSG00000130332 | LSM7 | 100 | 93.878 | ENSXETG00000022166 | lsm7 | 100 | 95.050 | Xenopus_tropicalis |
| ENSG00000130332 | LSM7 | 100 | 95.918 | ENSXCOG00000002572 | lsm7 | 100 | 95.098 | Xiphophorus_couchianus |
| ENSG00000130332 | LSM7 | 100 | 96.117 | ENSXMAG00000018630 | lsm7 | 100 | 96.117 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000398 | mRNA splicing, via spliceosome | 28781166. | IDA | Process |
| GO:0000398 | mRNA splicing, via spliceosome | - | TAS | Process |
| GO:0005515 | protein binding | 12165861.14603251.14667819.15231747.19628699.21988832.22365833.25416956. | IPI | Function |
| GO:0005634 | nucleus | 26912367. | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005688 | U6 snRNP | 21873635. | IBA | Component |
| GO:0005689 | U12-type spliceosomal complex | 21873635. | IBA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0017070 | U6 snRNA binding | 10523320. | NAS | Function |
| GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay | - | TAS | Process |
| GO:0046540 | U4/U6 x U5 tri-snRNP complex | 26912367. | IDA | Component |
| GO:0071004 | U2-type prespliceosome | 21873635. | IBA | Component |
| GO:0071005 | U2-type precatalytic spliceosome | 28781166. | IDA | Component |
| GO:0071013 | catalytic step 2 spliceosome | 21873635. | IBA | Component |
| GO:0120115 | Lsm2-8 complex | 10523320. | IDA | Component |
| GO:1990726 | Lsm1-7-Pat1 complex | 21873635. | IBA | Component |