
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| BRCA | |||
| LAML | |||
| LGG | |||
| LIHC | |||
| OV | |||
| THCA | |||
| THYM | |||
| UCEC | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| STAD | ||
| MESO | ||
| ACC | ||
| UCS | ||
| HNSC | ||
| PAAD | ||
| BLCA | ||
| CESC | ||
| LAML | ||
| UCEC | ||
| LIHC | ||
| DLBC | ||
| LGG | ||
| UVM |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000372393 | ASS1-203 | 1548 | XM_011518705 | ENSP00000361469 | 412 (aa) | XP_011517007 | P00966 |
| ENST00000422569 | ASS1-205 | 807 | - | ENSP00000394212 | 198 (aa) | - | Q5T6L6 |
| ENST00000352480 | ASS1-201 | 1564 | XM_005272200 | ENSP00000253004 | 412 (aa) | XP_005272257 | P00966 |
| ENST00000372386 | ASS1-202 | 667 | - | - | - (aa) | - | - |
| ENST00000443588 | ASS1-206 | 545 | - | ENSP00000397785 | 179 (aa) | - | Q5T6L5 |
| ENST00000372394 | ASS1-204 | 1973 | - | ENSP00000361471 | 412 (aa) | - | P00966 |
| ENST00000470849 | ASS1-208 | 652 | - | - | - (aa) | - | - |
| ENST00000492400 | ASS1-209 | 444 | - | - | - (aa) | - | - |
| ENST00000493984 | ASS1-210 | 687 | - | - | - (aa) | - | - |
| ENST00000467695 | ASS1-207 | 580 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000130707 | Lipoprotein(a) | 6.5240000E-005 | - |
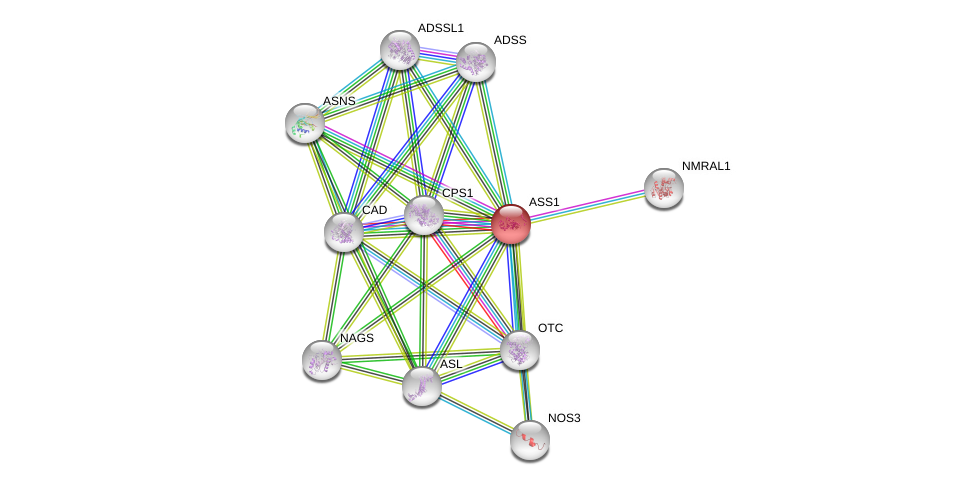
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000050 | urea cycle | 21873635. | IBA | Process |
| GO:0000050 | urea cycle | - | IEA | Process |
| GO:0000050 | urea cycle | 7977368.8792870.18473344.27287393. | IMP | Process |
| GO:0000050 | urea cycle | - | TAS | Process |
| GO:0000052 | citrulline metabolic process | 7977368. | IMP | Process |
| GO:0000053 | argininosuccinate metabolic process | 21873635. | IBA | Process |
| GO:0000053 | argininosuccinate metabolic process | 7977368. | IMP | Process |
| GO:0001822 | kidney development | - | IEA | Process |
| GO:0001889 | liver development | - | IEA | Process |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0004055 | argininosuccinate synthase activity | 21873635. | IBA | Function |
| GO:0004055 | argininosuccinate synthase activity | 7977368.8792870.18473344.27287393.28985504. | IMP | Function |
| GO:0005515 | protein binding | 12620389.17496144.28985504. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | - | IEA | Component |
| GO:0005737 | cytoplasm | 21873635. | IBA | Component |
| GO:0005741 | mitochondrial outer membrane | - | IEA | Component |
| GO:0005764 | lysosome | - | IEA | Component |
| GO:0005783 | endoplasmic reticulum | - | IEA | Component |
| GO:0005829 | cytosol | 28985504. | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0006526 | arginine biosynthetic process | 21873635. | IBA | Process |
| GO:0006526 | arginine biosynthetic process | - | IEA | Process |
| GO:0006526 | arginine biosynthetic process | 8792870.18473344.27287393. | IMP | Process |
| GO:0006531 | aspartate metabolic process | 7977368. | IMP | Process |
| GO:0006953 | acute-phase response | - | IEA | Process |
| GO:0007494 | midgut development | - | IEA | Process |
| GO:0007568 | aging | - | IEA | Process |
| GO:0007584 | response to nutrient | - | IEA | Process |
| GO:0007623 | circadian rhythm | 28985504. | IDA | Process |
| GO:0010043 | response to zinc ion | - | IEA | Process |
| GO:0010046 | response to mycotoxin | - | IEA | Process |
| GO:0015643 | toxic substance binding | - | IEA | Function |
| GO:0016597 | amino acid binding | 7977368. | IMP | Function |
| GO:0032355 | response to estradiol | - | IEA | Process |
| GO:0042802 | identical protein binding | 16189514.21988832.25416956. | IPI | Function |
| GO:0043204 | perikaryon | - | IEA | Component |
| GO:0045429 | positive regulation of nitric oxide biosynthetic process | 21106532. | IMP | Process |
| GO:0060416 | response to growth hormone | - | IEA | Process |
| GO:0060539 | diaphragm development | - | IEA | Process |
| GO:0070062 | extracellular exosome | 19056867.23533145. | HDA | Component |
| GO:0070852 | cell body fiber | - | IEA | Component |
| GO:0071222 | cellular response to lipopolysaccharide | - | IEA | Process |
| GO:0071230 | cellular response to amino acid stimulus | - | IEA | Process |
| GO:0071242 | cellular response to ammonium ion | - | IEA | Process |
| GO:0071320 | cellular response to cAMP | - | IEA | Process |
| GO:0071346 | cellular response to interferon-gamma | - | IEA | Process |
| GO:0071356 | cellular response to tumor necrosis factor | - | IEA | Process |
| GO:0071377 | cellular response to glucagon stimulus | - | IEA | Process |
| GO:0071400 | cellular response to oleic acid | - | IEA | Process |
| GO:0071418 | cellular response to amine stimulus | - | IEA | Process |
| GO:0071499 | cellular response to laminar fluid shear stress | 21106532. | IMP | Process |
| GO:0071549 | cellular response to dexamethasone stimulus | - | IEA | Process |
| GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 21106532. | IMP | Process |