
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| HNSC | |||
| KIRP | |||
| OV | |||
| PAAD | |||
| READ | |||
| SARC | |||
| TGCT | |||
| UCEC | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| STAD | ||
| MESO | ||
| ACC | ||
| SKCM | ||
| KIRP | ||
| PAAD | ||
| LIHC | ||
| DLBC | ||
| LGG | ||
| LUAD | ||
| UVM |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 29937225 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000595028 | TRIM28-205 | 486 | - | - | - (aa) | - | - |
| ENST00000597618 | TRIM28-210 | 609 | - | - | - (aa) | - | - |
| ENST00000597995 | TRIM28-212 | 702 | - | - | - (aa) | - | - |
| ENST00000253024 | TRIM28-201 | 2960 | XM_024451309 | ENSP00000253024 | 835 (aa) | XP_024307077 | Q13263 |
| ENST00000598355 | TRIM28-213 | 714 | - | - | - (aa) | - | - |
| ENST00000593582 | TRIM28-203 | 533 | - | ENSP00000472586 | 162 (aa) | - | M0R2I3 |
| ENST00000597136 | TRIM28-207 | 1552 | - | ENSP00000471303 | 460 (aa) | - | M0R0K9 |
| ENST00000594806 | TRIM28-204 | 861 | - | ENSP00000473126 | 178 (aa) | - | M0R3C0 |
| ENST00000597423 | TRIM28-209 | 733 | - | - | - (aa) | - | - |
| ENST00000595974 | TRIM28-206 | 665 | - | - | - (aa) | - | - |
| ENST00000597172 | TRIM28-208 | 577 | - | - | - (aa) | - | - |
| ENST00000597968 | TRIM28-211 | 504 | - | ENSP00000470503 | 61 (aa) | - | M0QZE6 |
| ENST00000601150 | TRIM28-215 | 915 | - | - | - (aa) | - | - |
| ENST00000341753 | TRIM28-202 | 2709 | - | ENSP00000342232 | 753 (aa) | - | Q13263 |
| ENST00000600840 | TRIM28-214 | 338 | - | - | - (aa) | - | - |
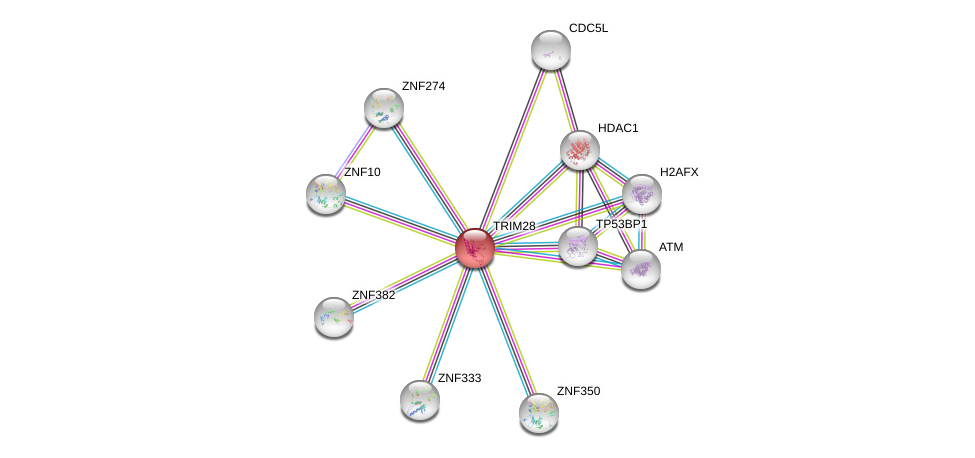
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000122 | negative regulation of transcription by RNA polymerase II | - | IEA | Process |
| GO:0000785 | chromatin | 21873635. | IBA | Component |
| GO:0001837 | epithelial to mesenchymal transition | - | ISS | Process |
| GO:0003677 | DNA binding | 21873635. | IBA | Function |
| GO:0003677 | DNA binding | 9016654. | IDA | Function |
| GO:0003682 | chromatin binding | 27029610. | IDA | Function |
| GO:0003713 | transcription coactivator activity | - | ISS | Function |
| GO:0003714 | transcription corepressor activity | 21873635. | IBA | Function |
| GO:0003714 | transcription corepressor activity | 8769649. | IDA | Function |
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0004672 | protein kinase activity | - | IEA | Function |
| GO:0004842 | ubiquitin-protein transferase activity | 21873635. | IBA | Function |
| GO:0004842 | ubiquitin-protein transferase activity | 18082607. | IDA | Function |
| GO:0005515 | protein binding | 10330177.11013263.16169070.17704056.18060868.18675275.18854139.19498464.20424263.20562864.20858735.20864041.20936779.21170338.21277283.21549307.21642969.21888893.23543754.23665872.24018422.24657165.26496610.27029610.27123980.27705803. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | 8769649.9016654. | IDA | Component |
| GO:0005634 | nucleus | - | ISS | Component |
| GO:0005654 | nucleoplasm | 21873635. | IBA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005654 | nucleoplasm | - | TAS | Component |
| GO:0005719 | nuclear euchromatin | - | IEA | Component |
| GO:0005720 | nuclear heterochromatin | - | IEA | Component |
| GO:0006281 | DNA repair | 17178852. | IDA | Process |
| GO:0006325 | chromatin organization | - | IEA | Process |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | - | TAS | Process |
| GO:0007265 | Ras protein signal transduction | 24623306. | IMP | Process |
| GO:0007566 | embryo implantation | - | IEA | Process |
| GO:0008270 | zinc ion binding | 10653693.11226167. | IDA | Function |
| GO:0016032 | viral process | - | IEA | Process |
| GO:0016567 | protein ubiquitination | - | IEA | Process |
| GO:0016925 | protein sumoylation | 21873635. | IBA | Process |
| GO:0016925 | protein sumoylation | 18082607. | IDA | Process |
| GO:0016925 | protein sumoylation | - | IEA | Process |
| GO:0019789 | SUMO transferase activity | 17298944. | EXP | Function |
| GO:0031625 | ubiquitin protein ligase binding | 18082607. | IDA | Function |
| GO:0035851 | Krueppel-associated box domain binding | 23665872. | IDA | Function |
| GO:0042307 | positive regulation of protein import into nucleus | 23665872. | IDA | Process |
| GO:0043045 | DNA methylation involved in embryo development | - | IEA | Process |
| GO:0043388 | positive regulation of DNA binding | - | IEA | Process |
| GO:0043565 | sequence-specific DNA binding | - | ISS | Function |
| GO:0045087 | innate immune response | 18248090. | IDA | Process |
| GO:0045739 | positive regulation of DNA repair | 17178852. | IDA | Process |
| GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate | - | IEA | Process |
| GO:0045892 | negative regulation of transcription, DNA-templated | 21873635. | IBA | Process |
| GO:0045892 | negative regulation of transcription, DNA-templated | 9016654. | IDA | Process |
| GO:0045893 | positive regulation of transcription, DNA-templated | - | ISS | Process |
| GO:0046777 | protein autophosphorylation | - | IEA | Process |
| GO:0051259 | protein complex oligomerization | 17512541. | IDA | Process |
| GO:0060028 | convergent extension involved in axis elongation | - | IEA | Process |
| GO:0060669 | embryonic placenta morphogenesis | - | IEA | Process |
| GO:0070087 | chromo shadow domain binding | 15882967. | IPI | Function |
| GO:0090309 | positive regulation of methylation-dependent chromatin silencing | 24623306. | IMP | Process |
| GO:0090309 | positive regulation of methylation-dependent chromatin silencing | - | ISS | Process |
| GO:0090575 | RNA polymerase II transcription factor complex | 21873635. | IBA | Component |
| GO:0090575 | RNA polymerase II transcription factor complex | - | ISS | Component |
| GO:1901536 | negative regulation of DNA demethylation | - | IEA | Process |
| GO:1902187 | negative regulation of viral release from host cell | 18248090. | IDA | Process |
| GO:1990841 | promoter-specific chromatin binding | 21873635. | IBA | Function |
| GO:1990841 | promoter-specific chromatin binding | 24623306. | IDA | Function |
| GO:2000653 | regulation of genetic imprinting | - | IEA | Process |