
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THCA | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| KIRP | |||
| KIRP | |||
| LAML | |||
| LUSC | |||
| MESO | |||
| PRAD | |||
| SKCM | |||
| THCA | |||
| THCA |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| ACC | ||
| UCS | ||
| SKCM | ||
| PRAD | ||
| LUSC | ||
| TGCT | ||
| COAD | ||
| PAAD | ||
| UCEC | ||
| LGG | ||
| LUAD |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000491754 | DIAPH1-209 | 557 | - | - | - (aa) | - | - |
| ENST00000494967 | DIAPH1-210 | 629 | - | - | - (aa) | - | - |
| ENST00000523100 | DIAPH1-213 | 1180 | - | ENSP00000428208 | 201 (aa) | - | B4E2I7 |
| ENST00000448451 | DIAPH1-205 | 2090 | - | ENSP00000408159 | 74 (aa) | - | H7C2W8 |
| ENST00000647330 | DIAPH1-217 | 1795 | - | ENSP00000494308 | 249 (aa) | - | A0A2R8YEF8 |
| ENST00000472516 | DIAPH1-207 | 508 | - | - | - (aa) | - | - |
| ENST00000643312 | DIAPH1-215 | 583 | - | ENSP00000495191 | 74 (aa) | - | H7C2W8 |
| ENST00000518484 | DIAPH1-212 | 546 | - | - | - (aa) | - | - |
| ENST00000476339 | DIAPH1-208 | 2606 | - | - | - (aa) | - | - |
| ENST00000468119 | DIAPH1-206 | 619 | - | ENSP00000493546 | 74 (aa) | - | H7C2W8 |
| ENST00000253811 | DIAPH1-201 | 5789 | - | ENSP00000253811 | 1228 (aa) | - | H9KV28 |
| ENST00000518047 | DIAPH1-211 | 4668 | XM_024454385 | ENSP00000428268 | 1263 (aa) | XP_024310153 | O60610 |
| ENST00000524301 | DIAPH1-214 | 561 | - | - | - (aa) | - | - |
| ENST00000389057 | DIAPH1-203 | 5762 | - | ENSP00000373709 | 1263 (aa) | - | A0A140T8Z0 |
| ENST00000389054 | DIAPH1-202 | 5795 | XM_024454384 | ENSP00000373706 | 1272 (aa) | XP_024310152 | O60610 |
| ENST00000643718 | DIAPH1-216 | 2228 | - | - | - (aa) | - | - |
| ENST00000398557 | DIAPH1-204 | 5789 | - | ENSP00000381565 | 1272 (aa) | - | A0A0G2JH68 |
| ENST00000647433 | DIAPH1-218 | 5843 | - | ENSP00000494675 | 1250 (aa) | - | A0A2R8Y5N1 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000131504 | rs72792324 | 5 | 141604894 | A | Mean platelet volume | 27863252 | [0.044-0.085] unit increase | 0.06435557 | EFO_0004584 |
| ENSG00000131504 | rs10066420 | 5 | 141594912 | A | Educational attainment (MTAG) | 30038396 | [0.0063-0.0117] unit decrease | 0.009 | EFO_0004784 |
| ENSG00000131504 | rs4912763 | 5 | 141592550 | T | Self-reported math ability (MTAG) | 30038396 | [0.0076-0.0158] unit decrease | 0.0117 | EFO_0004875 |
| ENSG00000131504 | rs6881581 | 5 | 141566788 | A | Educational attainment (years of education) | 30038396 | [0.0067-0.0125] unit decrease | 0.0096 | EFO_0004784 |
| ENSG00000131504 | rs10063055 | 5 | 141610541 | ? | Body mass index | 30595370 | EFO_0004340 | ||
| ENSG00000131504 | rs7712601 | 5 | 141538221 | ? | Eczema | 30595370 | HP_0000964 |
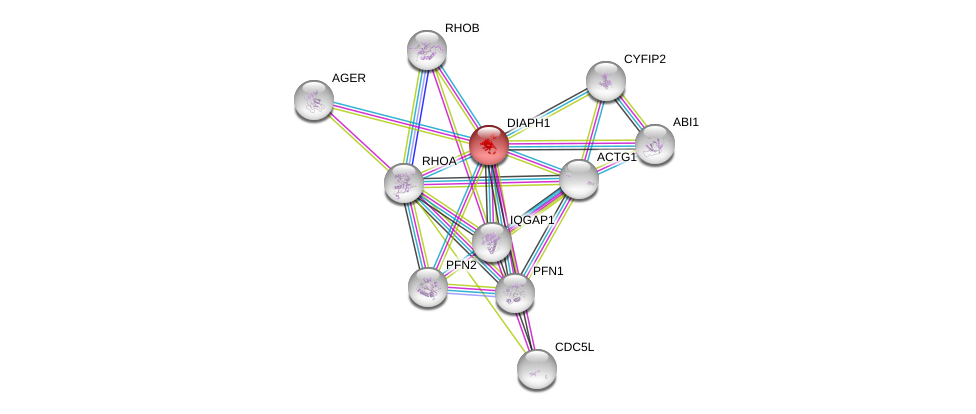
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0003779 | actin binding | - | IEA | Function |
| GO:0005102 | signaling receptor binding | 9360932. | NAS | Function |
| GO:0005515 | protein binding | 18218625.18230650.18922799.22190034. | IPI | Function |
| GO:0005634 | nucleus | - | ISS | Component |
| GO:0005737 | cytoplasm | - | ISS | Component |
| GO:0005815 | microtubule organizing center | - | IEA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005886 | plasma membrane | - | TAS | Component |
| GO:0007010 | cytoskeleton organization | 21834987. | IMP | Process |
| GO:0007010 | cytoskeleton organization | - | ISS | Process |
| GO:0007605 | sensory perception of sound | - | IEA | Process |
| GO:0008360 | regulation of cell shape | 21834987. | IMP | Process |
| GO:0017048 | Rho GTPase binding | - | IEA | Function |
| GO:0030036 | actin cytoskeleton organization | - | ISS | Process |
| GO:0030041 | actin filament polymerization | - | ISS | Process |
| GO:0030335 | positive regulation of cell migration | - | IEA | Process |
| GO:0030667 | secretory granule membrane | - | TAS | Component |
| GO:0032587 | ruffle membrane | - | IEA | Component |
| GO:0032886 | regulation of microtubule-based process | 20937854. | IMP | Process |
| GO:0035372 | protein localization to microtubule | 15123714. | IMP | Process |
| GO:0043312 | neutrophil degranulation | - | TAS | Process |
| GO:0044325 | ion channel binding | 15123714. | IPI | Function |
| GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 15123714. | IMP | Process |
| GO:0051493 | regulation of cytoskeleton organization | 26912466. | IMP | Process |
| GO:0071420 | cellular response to histamine | 15123714. | IMP | Process |
| GO:0072686 | mitotic spindle | 15123714. | IDA | Component |
| GO:0101003 | ficolin-1-rich granule membrane | - | TAS | Component |
| GO:2000145 | regulation of cell motility | - | TAS | Process |