
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| DLBC | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| MESO | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| DLBC | |||
| TGCT |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| STAD | ||
| SARC | ||
| MESO | ||
| ACC | ||
| PRAD | ||
| KIRP | ||
| CESC | ||
| READ | ||
| UCEC | ||
| LIHC | ||
| DLBC | ||
| LGG | ||
| UVM | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000350030 | NASP-201 | 3207 | XM_005270888 | ENSP00000255120 | 788 (aa) | XP_005270945 | P49321 |
| ENST00000530073 | NASP-218 | 2740 | - | - | - (aa) | - | - |
| ENST00000531532 | NASP-220 | 554 | - | ENSP00000436778 | 56 (aa) | - | E9PKR5 |
| ENST00000351223 | NASP-202 | 1538 | XM_005270889 | ENSP00000255121 | 449 (aa) | XP_005270946 | P49321 |
| ENST00000453748 | NASP-206 | 2080 | - | ENSP00000403157 | 38 (aa) | - | E9PAU3 |
| ENST00000528084 | NASP-215 | 535 | - | ENSP00000435363 | 56 (aa) | - | E9PKR5 |
| ENST00000529333 | NASP-217 | 778 | - | ENSP00000436388 | 38 (aa) | - | E9PAU3 |
| ENST00000437901 | NASP-205 | 874 | - | ENSP00000400792 | 256 (aa) | - | E9PI86 |
| ENST00000629893 | NASP-225 | 253 | - | ENSP00000486336 | 56 (aa) | - | E9PKR5 |
| ENST00000537798 | NASP-224 | 3097 | - | ENSP00000438871 | 724 (aa) | - | P49321 |
| ENST00000534101 | NASP-222 | 754 | - | - | - (aa) | - | - |
| ENST00000527359 | NASP-212 | 743 | - | ENSP00000436790 | 37 (aa) | - | E9PQG5 |
| ENST00000531612 | NASP-221 | 938 | - | ENSP00000437116 | 288 (aa) | - | H0YF33 |
| ENST00000481782 | NASP-210 | 3659 | - | - | - (aa) | - | - |
| ENST00000472408 | NASP-209 | 541 | - | - | - (aa) | - | - |
| ENST00000437362 | NASP-204 | 999 | - | ENSP00000388010 | 38 (aa) | - | E9PAU3 |
| ENST00000527932 | NASP-214 | 650 | - | - | - (aa) | - | - |
| ENST00000470768 | NASP-208 | 1036 | - | ENSP00000436924 | 316 (aa) | - | E9PPR5 |
| ENST00000527470 | NASP-213 | 532 | - | ENSP00000437241 | 143 (aa) | - | E9PNB5 |
| ENST00000528238 | NASP-216 | 894 | - | ENSP00000432289 | 268 (aa) | - | E9PRH9 |
| ENST00000530840 | NASP-219 | 531 | - | ENSP00000435548 | 55 (aa) | - | E9PJQ2 |
| ENST00000372052 | NASP-203 | 1458 | - | ENSP00000361122 | 422 (aa) | - | Q5T624 |
| ENST00000534450 | NASP-223 | 632 | - | ENSP00000434240 | 175 (aa) | - | H0YDS9 |
| ENST00000525515 | NASP-211 | 565 | - | ENSP00000436939 | 154 (aa) | - | E9PPQ8 |
| ENST00000464190 | NASP-207 | 604 | - | - | - (aa) | - | - |
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| ENSG00000132780 | Metabolism | 5E-11 | 24816252 |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000132780 | rs1053941 | 1 | 45618711 | T | Blood metabolite levels | 24816252 | [0.011-0.023] unit decrease | 0.017 | EFO_0005664 |
| ENSG00000132780 | rs72688441 | 1 | 45585381 | A | Blood protein levels | 29875488 | [1.08-1.28] unit decrease | 1.18 | EFO_0007937 |
| ENSG00000132780 | rs2230657 | 1 | 45607817 | A | Hemoglobin concentration | 27863252 | [0.019-0.033] unit decrease | 0.02592878 | EFO_0004509 |
| ENSG00000132780 | rs72688441 | 1 | 45585381 | G | Blood protein levels | 30072576 | [1.18-1.35] unit increase | 1.267 | EFO_0007937 |
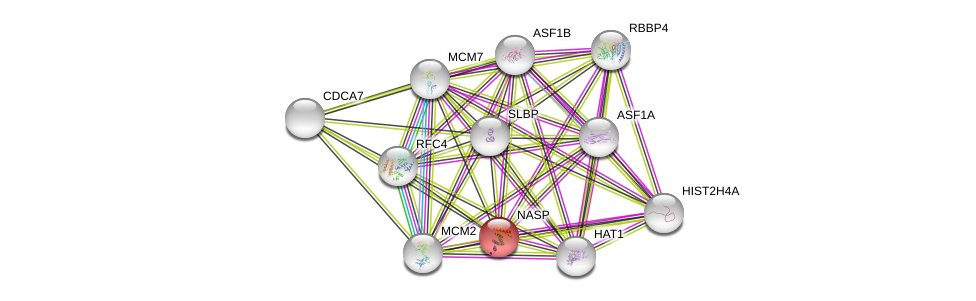
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000132780 | NASP | 95 | 97.561 | ENSMUSG00000028693 | Nasp | 99 | 95.620 | Mus_musculus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000082 | G1/S transition of mitotic cell cycle | - | ISS | Process |
| GO:0000790 | nuclear chromatin | 14718166. | IDA | Component |
| GO:0001824 | blastocyst development | - | ISS | Process |
| GO:0005515 | protein binding | 16189514.19410544.20080577.21044950.29568061. | IPI | Function |
| GO:0005634 | nucleus | 21873635. | IBA | Component |
| GO:0005634 | nucleus | - | ISS | Component |
| GO:0005654 | nucleoplasm | 21873635. | IBA | Component |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005737 | cytoplasm | - | IEA | Component |
| GO:0006260 | DNA replication | - | ISS | Process |
| GO:0006335 | DNA replication-dependent nucleosome assembly | 21873635. | IBA | Process |
| GO:0006335 | DNA replication-dependent nucleosome assembly | 14718166. | IDA | Process |
| GO:0006336 | DNA replication-independent nucleosome assembly | 14718166. | IDA | Process |
| GO:0008283 | cell proliferation | - | ISS | Process |
| GO:0008584 | male gonad development | - | IEA | Process |
| GO:0015031 | protein transport | - | IEA | Process |
| GO:0032991 | protein-containing complex | 14718166. | IDA | Component |
| GO:0033574 | response to testosterone | - | IEA | Process |
| GO:0034080 | CENP-A containing nucleosome assembly | 21873635. | IBA | Process |
| GO:0042393 | histone binding | 21873635. | IBA | Function |
| GO:0042393 | histone binding | 27618665. | IDA | Function |
| GO:0043486 | histone exchange | - | ISS | Process |