
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CHOL | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| KIRP | |||||||
| LAML | |||||||
| LAML | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PCPG | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| TGCT | |||||||
| THYM | |||||||
| THYM | |||||||
| THYM | |||||||
| THYM | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCS | |||||||
| UCS | |||||||
| UCS | |||||||
| UCS |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CESC | |||
| KIRP | |||
| LUSC | |||
| OV | |||
| PAAD | |||
| TGCT | |||
| UCEC |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| ACC | ||
| SKCM | ||
| KIRP | ||
| COAD | ||
| PAAD | ||
| CESC | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| THCA |
| PID | Title | Article | Time | Author | Doi |
|---|---|---|---|---|---|
| 25087874 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000462366 | ROCK2-206 | 563 | - | - | - (aa) | - | - |
| ENST00000261535 | ROCK2-201 | 1795 | - | ENSP00000261535 | 446 (aa) | - | D6REE7 |
| ENST00000401753 | ROCK2-203 | 4017 | - | ENSP00000385509 | 1145 (aa) | - | E9PF63 |
| ENST00000484951 | ROCK2-207 | 895 | - | - | - (aa) | - | - |
| ENST00000493096 | ROCK2-208 | 461 | - | - | - (aa) | - | - |
| ENST00000460262 | ROCK2-205 | 597 | - | - | - (aa) | - | - |
| ENST00000616279 | ROCK2-209 | 6401 | - | ENSP00000481789 | 703 (aa) | - | Q14DU5 |
| ENST00000315872 | ROCK2-202 | 8292 | XM_005246190 | ENSP00000317985 | 1388 (aa) | XP_005246247 | O75116 |
| ENST00000431087 | ROCK2-204 | 729 | - | ENSP00000395957 | 203 (aa) | - | C9JFJ0 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04022 | cGMP-PKG signaling pathway | KEGG |
| hsa04024 | cAMP signaling pathway | KEGG |
| hsa04062 | Chemokine signaling pathway | KEGG |
| hsa04071 | Sphingolipid signaling pathway | KEGG |
| hsa04270 | Vascular smooth muscle contraction | KEGG |
| hsa04310 | Wnt signaling pathway | KEGG |
| hsa04360 | Axon guidance | KEGG |
| hsa04510 | Focal adhesion | KEGG |
| hsa04530 | Tight junction | KEGG |
| hsa04611 | Platelet activation | KEGG |
| hsa04670 | Leukocyte transendothelial migration | KEGG |
| hsa04810 | Regulation of actin cytoskeleton | KEGG |
| hsa04921 | Oxytocin signaling pathway | KEGG |
| hsa05130 | Pathogenic Escherichia coli infection | KEGG |
| hsa05131 | Shigellosis | KEGG |
| hsa05132 | Salmonella infection | KEGG |
| hsa05163 | Human cytomegalovirus infection | KEGG |
| hsa05200 | Pathways in cancer | KEGG |
| hsa05205 | Proteoglycans in cancer | KEGG |
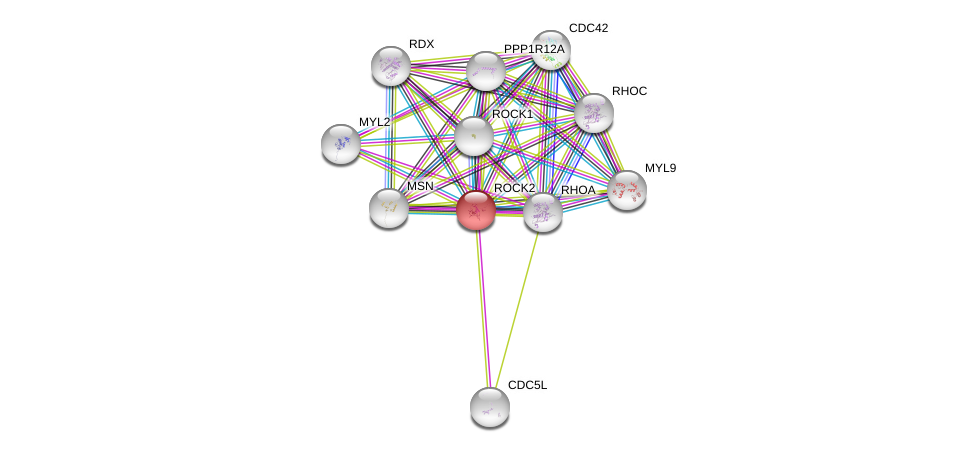
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000134318 | ROCK2 | 100 | 94.118 | ENSG00000067900 | ROCK1 | 99 | 65.132 | Homo_sapiens |
| ENSG00000134318 | ROCK2 | 98 | 52.206 | ENSG00000198752 | CDC42BPB | 88 | 50.877 | Homo_sapiens |
| ENSG00000134318 | ROCK2 | 99 | 42.424 | ENSG00000122966 | CIT | 90 | 40.455 | Homo_sapiens |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0001934 | positive regulation of protein phosphorylation | 24065547. | IMP | Process |
| GO:0002931 | response to ischemia | - | ISS | Process |
| GO:0003180 | aortic valve morphogenesis | - | ISS | Process |
| GO:0003723 | RNA binding | 22681889. | HDA | Function |
| GO:0004674 | protein serine/threonine kinase activity | 16574662.21147781. | IDA | Function |
| GO:0004674 | protein serine/threonine kinase activity | 24305806.29353861. | IMP | Function |
| GO:0004674 | protein serine/threonine kinase activity | 24305806. | ISS | Function |
| GO:0004674 | protein serine/threonine kinase activity | 27853422. | NAS | Function |
| GO:0005198 | structural molecule activity | 9933571. | NAS | Function |
| GO:0005515 | protein binding | 17015463.20230755. | IPI | Function |
| GO:0005524 | ATP binding | - | IEA | Function |
| GO:0005634 | nucleus | - | IEA | Component |
| GO:0005813 | centrosome | 17015463. | IDA | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005886 | plasma membrane | - | IEA | Component |
| GO:0006468 | protein phosphorylation | 18559669. | IDA | Process |
| GO:0006939 | smooth muscle contraction | 12778124. | TAS | Process |
| GO:0007186 | G protein-coupled receptor signaling pathway | - | TAS | Process |
| GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 25816133. | IMP | Process |
| GO:0007266 | Rho protein signal transduction | - | IEA | Process |
| GO:0010595 | positive regulation of endothelial cell migration | 24065547. | IMP | Process |
| GO:0010613 | positive regulation of cardiac muscle hypertrophy | - | ISS | Process |
| GO:0010628 | positive regulation of gene expression | 24065547. | IMP | Process |
| GO:0010628 | positive regulation of gene expression | - | ISS | Process |
| GO:0010629 | negative regulation of gene expression | - | ISS | Process |
| GO:0010825 | positive regulation of centrosome duplication | - | IEA | Process |
| GO:0016525 | negative regulation of angiogenesis | 19181962. | IMP | Process |
| GO:0017048 | Rho GTPase binding | - | IEA | Function |
| GO:0018105 | peptidyl-serine phosphorylation | 24305806. | IMP | Process |
| GO:0018105 | peptidyl-serine phosphorylation | 24305806. | ISS | Process |
| GO:0018107 | peptidyl-threonine phosphorylation | 24305806.29353861. | IMP | Process |
| GO:0018107 | peptidyl-threonine phosphorylation | - | ISS | Process |
| GO:0030155 | regulation of cell adhesion | 12778124. | TAS | Process |
| GO:0030335 | positive regulation of cell migration | 20232393. | IMP | Process |
| GO:0030866 | cortical actin cytoskeleton organization | 25816133. | IGI | Process |
| GO:0030866 | cortical actin cytoskeleton organization | 25816133. | IMP | Process |
| GO:0031644 | regulation of neurological system process | - | ISS | Process |
| GO:0032723 | positive regulation of connective tissue growth factor production | - | ISS | Process |
| GO:0032956 | regulation of actin cytoskeleton organization | 12778124. | TAS | Process |
| GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity | 25816133. | IGI | Process |
| GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity | 25816133. | IMP | Process |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 15121898. | IDA | Component |
| GO:0039694 | viral RNA genome replication | 19376974. | IMP | Process |
| GO:0042752 | regulation of circadian rhythm | - | ISS | Process |
| GO:0043410 | positive regulation of MAPK cascade | - | ISS | Process |
| GO:0045019 | negative regulation of nitric oxide biosynthetic process | 29353861. | IMP | Process |
| GO:0045019 | negative regulation of nitric oxide biosynthetic process | - | ISS | Process |
| GO:0045616 | regulation of keratinocyte differentiation | 19997641. | IMP | Process |
| GO:0046872 | metal ion binding | - | IEA | Function |
| GO:0048010 | vascular endothelial growth factor receptor signaling pathway | - | TAS | Process |
| GO:0048013 | ephrin receptor signaling pathway | - | TAS | Process |
| GO:0048156 | tau protein binding | 28386764. | NAS | Function |
| GO:0048511 | rhythmic process | - | IEA | Process |
| GO:0050321 | tau-protein kinase activity | 28386764. | NAS | Function |
| GO:0051246 | regulation of protein metabolic process | 24305806. | ISS | Process |
| GO:0051298 | centrosome duplication | 17015463. | IMP | Process |
| GO:0051492 | regulation of stress fiber assembly | 12778124. | TAS | Process |
| GO:0051496 | positive regulation of stress fiber assembly | 20232393. | IMP | Process |
| GO:0051893 | regulation of focal adhesion assembly | 12778124. | TAS | Process |
| GO:0061157 | mRNA destabilization | - | ISS | Process |
| GO:0070168 | negative regulation of biomineral tissue development | - | ISS | Process |
| GO:0071394 | cellular response to testosterone stimulus | 24065547. | IMP | Process |
| GO:0071559 | response to transforming growth factor beta | - | ISS | Process |
| GO:0072518 | Rho-dependent protein serine/threonine kinase activity | 24065547. | IMP | Function |
| GO:0072518 | Rho-dependent protein serine/threonine kinase activity | 28679962. | TAS | Function |
| GO:0072659 | protein localization to plasma membrane | 25816133. | IGI | Process |
| GO:0072659 | protein localization to plasma membrane | 25816133. | IMP | Process |
| GO:0090271 | positive regulation of fibroblast growth factor production | - | ISS | Process |
| GO:0097746 | regulation of blood vessel diameter | - | ISS | Process |
| GO:0110061 | regulation of angiotensin-activated signaling pathway | - | ISS | Process |
| GO:0150033 | negative regulation of protein localization to lysosome | 24305806. | IMP | Process |
| GO:1900037 | regulation of cellular response to hypoxia | - | ISS | Process |
| GO:1902004 | positive regulation of amyloid-beta formation | 24305806. | IMP | Process |
| GO:1902004 | positive regulation of amyloid-beta formation | 24305806. | ISS | Process |
| GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process | 24305806. | IMP | Process |
| GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process | 24305806. | ISS | Process |
| GO:1902966 | positive regulation of protein localization to early endosome | 24305806. | IMP | Process |
| GO:1902993 | positive regulation of amyloid precursor protein catabolic process | 24305806. | IMP | Process |
| GO:1902993 | positive regulation of amyloid precursor protein catabolic process | 24305806. | ISS | Process |
| GO:1903140 | regulation of establishment of endothelial barrier | 25816133. | IGI | Process |
| GO:1903140 | regulation of establishment of endothelial barrier | 25816133. | IMP | Process |
| GO:1903347 | negative regulation of bicellular tight junction assembly | 25816133. | IGI | Process |
| GO:1905145 | cellular response to acetylcholine | - | ISS | Process |
| GO:1905205 | positive regulation of connective tissue replacement | - | ISS | Process |
| GO:1990776 | response to angiotensin | - | ISS | Process |
| GO:2000114 | regulation of establishment of cell polarity | 20803696. | TAS | Process |
| GO:2000145 | regulation of cell motility | 12778124. | TAS | Process |