
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| GBM | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| LGG | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| COAD | |||
| DLBC | |||
| GBM | |||
| LUAD | |||
| MESO | |||
| PAAD | |||
| READ | |||
| STAD | |||
| THYM |
| Cancer | P-value | Q-value |
|---|---|---|
| KIRC | ||
| STAD | ||
| SARC | ||
| MESO | ||
| ACC | ||
| HNSC | ||
| PAAD | ||
| BLCA | ||
| LAML | ||
| UCEC | ||
| LUAD | ||
| UVM |
| Protein ID | Domain | Pfam ID | E-value | Domain number | Total number |
|---|---|---|---|---|---|
| ENSP00000256953 | MMR_HSR1 | PF01926.23 | 0.00017 | 1 | 1 |
| ENSP00000438280 | MMR_HSR1 | PF01926.23 | 0.00017 | 1 | 1 |
| ENSP00000441505 | MMR_HSR1 | PF01926.23 | 0.00017 | 1 | 1 |
| ENSP00000441860 | MMR_HSR1 | PF01926.23 | 0.00075 | 1 | 1 |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000256953 | RERG-201 | 2264 | XM_017020121 | ENSP00000256953 | 199 (aa) | XP_016875610 | Q96A58 |
| ENST00000393736 | RERG-202 | 655 | - | ENSP00000440887 | 38 (aa) | - | F5GYR1 |
| ENST00000437578 | RERG-203 | 562 | - | - | - (aa) | - | - |
| ENST00000538313 | RERG-207 | 2060 | - | ENSP00000441505 | 199 (aa) | - | Q96A58 |
| ENST00000537647 | RERG-205 | 935 | - | ENSP00000441860 | 70 (aa) | - | F5GWE9 |
| ENST00000545567 | RERG-208 | 574 | - | ENSP00000439532 | 113 (aa) | - | F5H252 |
| ENST00000536465 | RERG-204 | 2134 | - | ENSP00000438280 | 199 (aa) | - | Q96A58 |
| ENST00000546331 | RERG-209 | 1383 | - | ENSP00000444485 | 180 (aa) | - | Q96A58 |
| ENST00000537717 | RERG-206 | 582 | - | - | - (aa) | - | - |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000134533 | rs12311304 | 12 | 15236703 | G | Behavioural disinhibition (generation interaction) | 23942779 | EFO_0005430|EFO_0008364|EFO_0005431|EFO_0005432|EFO_0004329|EFO_0003829 | ||
| ENSG00000134533 | rs7956634 | 12 | 15168260 | T | Glomerular filtration rate (creatinine) | 26831199 | [0.0048-0.0088] unit decrease | 0.0068 | EFO_0005208|EFO_0004518 |
| ENSG00000134533 | rs12300553 | 12 | 15228276 | G | Urate levels in overweight individuals | 25811787 | [0.045-0.115] kg/m2 decrease | 0.08 | EFO_0004531|EFO_0005935 |
| ENSG00000134533 | rs1117152 | 12 | 15285098 | T | Educational attainment (MTAG) | 30038396 | [0.0076-0.013] unit increase | 0.0103 | EFO_0004784 |
| ENSG00000134533 | rs12826808 | 12 | 15170446 | ? | Glomerular filtration rate (creatinine) | 28452372 | EFO_0005208|EFO_0004518 | ||
| ENSG00000134533 | rs1117152 | 12 | 15285098 | T | Educational attainment (years of education) | 30038396 | [0.0077-0.0135] unit increase | 0.0106 | EFO_0004784 |
| ENSG00000134533 | rs11056375 | 12 | 15154051 | ? | Red blood cell count | 30595370 | EFO_0004305 | ||
| ENSG00000134533 | rs7313556 | 12 | 15144425 | A | Diastolic blood pressure | 30224653 | [0.059-0.121] mmHg increase | 0.09 | EFO_0006336 |
| ENSG00000134533 | rs7306438 | 12 | 15124179 | ? | Lung function (FEV1/FVC) | 30595370 | EFO_0004713 |
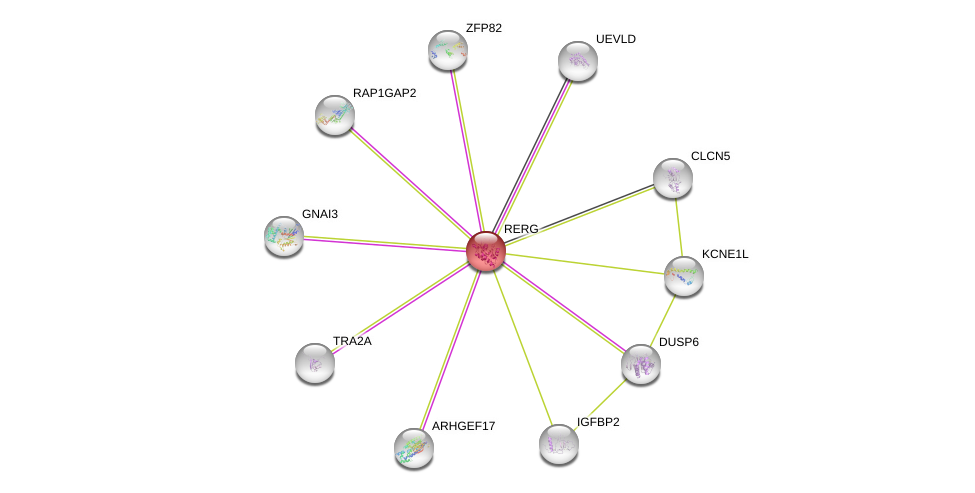
| Ensembl ID | Gene Symbol | Coverage | Identiy | Ortholog | Gene Symbol | Coverage | Identiy | Species |
|---|---|---|---|---|---|---|---|---|
| ENSG00000134533 | RERG | 100 | 92.105 | ENSAPOG00000015680 | - | 100 | 82.000 | Acanthochromis_polyacanthus |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSAOCG00000010160 | si:cabz01085950.1 | 100 | 82.500 | Amphiprion_ocellaris |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSAPEG00000018205 | si:cabz01085950.1 | 100 | 82.500 | Amphiprion_percula |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSATEG00000004893 | si:cabz01085950.1 | 100 | 81.500 | Anabas_testudineus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSANAG00000034086 | RERG | 100 | 99.497 | Aotus_nancymaae |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSACLG00000009103 | si:cabz01085950.1 | 100 | 83.000 | Astatotilapia_calliptera |
| ENSG00000134533 | RERG | 100 | 94.737 | ENSAMXG00000033069 | si:cabz01085950.1 | 100 | 80.296 | Astyanax_mexicanus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSBTAG00000002174 | RERG | 100 | 99.497 | Bos_taurus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSCJAG00000021602 | RERG | 100 | 98.995 | Callithrix_jacchus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSCAFG00000012854 | RERG | 100 | 99.497 | Canis_familiaris |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSCAFG00020007046 | RERG | 100 | 99.497 | Canis_lupus_dingo |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSCHIG00000011004 | RERG | 100 | 99.497 | Capra_hircus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSTSYG00000011437 | RERG | 100 | 98.995 | Carlito_syrichta |
| ENSG00000134533 | RERG | 100 | 98.995 | ENSCPOG00000000528 | RERG | 100 | 98.995 | Cavia_porcellus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSCCAG00000026139 | RERG | 100 | 98.995 | Cebus_capucinus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSCATG00000030178 | RERG | 100 | 100.000 | Cercocebus_atys |
| ENSG00000134533 | RERG | 100 | 99.497 | ENSCLAG00000016162 | RERG | 100 | 99.497 | Chinchilla_lanigera |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSCSAG00000008360 | RERG | 100 | 100.000 | Chlorocebus_sabaeus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSCPBG00000028244 | RERG | 100 | 96.482 | Chrysemys_picta_bellii |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSCANG00000040826 | RERG | 100 | 100.000 | Colobus_angolensis_palliatus |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSCSEG00000015770 | si:cabz01085950.1 | 100 | 82.000 | Cynoglossus_semilaevis |
| ENSG00000134533 | RERG | 100 | 89.474 | ENSCVAG00000006083 | si:cabz01085950.1 | 100 | 81.500 | Cyprinodon_variegatus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSDNOG00000043703 | RERG | 100 | 99.497 | Dasypus_novemcinctus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSETEG00000010802 | RERG | 100 | 95.980 | Echinops_telfairi |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSEASG00005014613 | RERG | 100 | 99.497 | Equus_asinus_asinus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSECAG00000014358 | RERG | 100 | 99.497 | Equus_caballus |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSELUG00000013666 | si:cabz01085950.1 | 100 | 84.000 | Esox_lucius |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSFCAG00000029875 | RERG | 100 | 98.492 | Felis_catus |
| ENSG00000134533 | RERG | 100 | 98.000 | ENSFDAG00000007662 | RERG | 100 | 98.000 | Fukomys_damarensis |
| ENSG00000134533 | RERG | 100 | 89.474 | ENSFHEG00000016504 | si:cabz01085950.1 | 100 | 81.500 | Fundulus_heteroclitus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSGALG00000028186 | RERG | 100 | 95.980 | Gallus_gallus |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSGACG00000019134 | si:cabz01085950.1 | 100 | 81.000 | Gasterosteus_aculeatus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSGAGG00000021838 | RERG | 100 | 95.980 | Gopherus_agassizii |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSHBUG00000011397 | si:cabz01085950.1 | 100 | 83.000 | Haplochromis_burtoni |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSHGLG00000018663 | RERG | 100 | 100.000 | Heterocephalus_glaber_female |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSHGLG00100003290 | RERG | 100 | 100.000 | Heterocephalus_glaber_male |
| ENSG00000134533 | RERG | 100 | 86.842 | ENSIPUG00000005254 | RERG | 100 | 78.325 | Ictalurus_punctatus |
| ENSG00000134533 | RERG | 100 | 99.497 | ENSSTOG00000023505 | RERG | 100 | 99.497 | Ictidomys_tridecemlineatus |
| ENSG00000134533 | RERG | 100 | 89.474 | ENSKMAG00000003080 | si:cabz01085950.1 | 100 | 80.000 | Kryptolebias_marmoratus |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSLBEG00000027628 | si:cabz01085950.1 | 100 | 82.500 | Labrus_bergylta |
| ENSG00000134533 | RERG | 100 | 97.368 | ENSLOCG00000016464 | si:cabz01085950.1 | 100 | 91.457 | Lepisosteus_oculatus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSLAFG00000001217 | RERG | 100 | 100.000 | Loxodonta_africana |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSMFAG00000041631 | RERG | 100 | 100.000 | Macaca_fascicularis |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSMMUG00000010460 | RERG | 100 | 100.000 | Macaca_mulatta |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSMNEG00000042410 | RERG | 100 | 100.000 | Macaca_nemestrina |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSMLEG00000043731 | RERG | 100 | 100.000 | Mandrillus_leucophaeus |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSMAMG00000001320 | si:cabz01085950.1 | 85 | 81.500 | Mastacembelus_armatus |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSMZEG00005018287 | si:cabz01085950.1 | 100 | 83.000 | Maylandia_zebra |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSMGAG00000013542 | RERG | 100 | 95.980 | Meleagris_gallopavo |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSMICG00000006266 | RERG | 100 | 98.995 | Microcebus_murinus |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSMMOG00000006335 | si:cabz01085950.1 | 100 | 82.500 | Mola_mola |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSMPUG00000011559 | RERG | 100 | 99.497 | Mustela_putorius_furo |
| ENSG00000134533 | RERG | 100 | 97.487 | ENSNGAG00000000037 | Rerg | 100 | 97.487 | Nannospalax_galili |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSNLEG00000029492 | RERG | 100 | 84.080 | Nomascus_leucogenys |
| ENSG00000134533 | RERG | 100 | 97.990 | ENSOPRG00000015124 | RERG | 100 | 97.990 | Ochotona_princeps |
| ENSG00000134533 | RERG | 100 | 98.995 | ENSODEG00000009994 | RERG | 100 | 98.995 | Octodon_degus |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSONIG00000009978 | si:cabz01085950.1 | 100 | 83.000 | Oreochromis_niloticus |
| ENSG00000134533 | RERG | 100 | 97.990 | ENSOCUG00000000650 | RERG | 100 | 97.990 | Oryctolagus_cuniculus |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSORLG00020005258 | rerg | 100 | 82.000 | Oryzias_latipes_hni |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSORLG00015002284 | rerg | 100 | 82.000 | Oryzias_latipes_hsok |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSOARG00000020676 | RERG | 100 | 99.497 | Ovis_aries |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSPPAG00000004422 | RERG | 100 | 99.497 | Pan_paniscus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSPPRG00000003131 | RERG | 100 | 98.492 | Panthera_pardus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSPTIG00000008594 | RERG | 100 | 98.492 | Panthera_tigris_altaica |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSPTRG00000004731 | RERG | 100 | 99.497 | Pan_troglodytes |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSPANG00000005456 | RERG | 100 | 100.000 | Papio_anubis |
| ENSG00000134533 | RERG | 100 | 94.737 | ENSPKIG00000002719 | si:cabz01085950.1 | 100 | 84.925 | Paramormyrops_kingsleyae |
| ENSG00000134533 | RERG | 100 | 97.368 | ENSPSIG00000003221 | RERG | 100 | 95.477 | Pelodiscus_sinensis |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSPMGG00000004104 | si:cabz01085950.1 | 100 | 81.500 | Periophthalmus_magnuspinnatus |
| ENSG00000134533 | RERG | 100 | 86.842 | ENSPMAG00000006671 | si:cabz01085950.1 | 100 | 71.642 | Petromyzon_marinus |
| ENSG00000134533 | RERG | 100 | 89.474 | ENSPFOG00000008678 | si:cabz01085950.1 | 100 | 81.500 | Poecilia_formosa |
| ENSG00000134533 | RERG | 100 | 89.474 | ENSPMEG00000011210 | si:cabz01085950.1 | 100 | 81.500 | Poecilia_mexicana |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSPPYG00000004331 | RERG | 100 | 100.000 | Pongo_abelii |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSPVAG00000013319 | RERG | 100 | 99.497 | Pteropus_vampyrus |
| ENSG00000134533 | RERG | 100 | 94.737 | ENSPNAG00000028497 | si:cabz01085950.1 | 100 | 81.281 | Pygocentrus_nattereri |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSRBIG00000036581 | RERG | 100 | 100.000 | Rhinopithecus_bieti |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSRROG00000042897 | RERG | 100 | 100.000 | Rhinopithecus_roxellana |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSSBOG00000034195 | RERG | 100 | 98.492 | Saimiri_boliviensis_boliviensis |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSSMAG00000014329 | si:cabz01085950.1 | 100 | 82.000 | Scophthalmus_maximus |
| ENSG00000134533 | RERG | 100 | 89.474 | ENSSDUG00000014082 | si:cabz01085950.1 | 100 | 82.000 | Seriola_dumerili |
| ENSG00000134533 | RERG | 100 | 89.474 | ENSSLDG00000013333 | si:cabz01085950.1 | 100 | 82.500 | Seriola_lalandi_dorsalis |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSSPAG00000018992 | si:cabz01085950.1 | 100 | 82.500 | Stegastes_partitus |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSTRUG00000023011 | rerg | 100 | 82.000 | Takifugu_rubripes |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSTNIG00000006917 | si:cabz01085950.1 | 100 | 83.846 | Tetraodon_nigroviridis |
| ENSG00000134533 | RERG | 100 | 98.492 | ENSTTRG00000015290 | RERG | 100 | 98.492 | Tursiops_truncatus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSUAMG00000010578 | RERG | 100 | 98.995 | Ursus_americanus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSUMAG00000002671 | RERG | 100 | 98.995 | Ursus_maritimus |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSVPAG00000002449 | RERG | 100 | 98.492 | Vicugna_pacos |
| ENSG00000134533 | RERG | 100 | 100.000 | ENSVVUG00000004380 | RERG | 100 | 99.497 | Vulpes_vulpes |
| ENSG00000134533 | RERG | 100 | 92.105 | ENSXETG00000022517 | rerg | 100 | 89.447 | Xenopus_tropicalis |
| ENSG00000134533 | RERG | 100 | 89.474 | ENSXMAG00000024024 | si:cabz01085950.1 | 100 | 81.500 | Xiphophorus_maculatus |
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0003924 | GTPase activity | 11533059. | IDA | Function |
| GO:0005525 | GTP binding | - | IEA | Function |
| GO:0005634 | nucleus | 11533059. | IDA | Component |
| GO:0005829 | cytosol | 11533059. | IDA | Component |
| GO:0007264 | small GTPase mediated signal transduction | 11533059. | NAS | Process |
| GO:0008285 | negative regulation of cell proliferation | 11533059. | IDA | Process |
| GO:0009725 | response to hormone | 11533059. | IDA | Process |
| GO:0016020 | membrane | - | IEA | Component |
| GO:0019003 | GDP binding | 11533059. | TAS | Function |
| GO:0030308 | negative regulation of cell growth | 11533059. | IDA | Process |
| GO:0030331 | estrogen receptor binding | 11533059. | NAS | Function |