
| Cancer | Chr | Position | Mutation Type | dbSNP | Protein-change | Allele Freq | RBD |
|---|---|---|---|---|---|---|---|
| ACC | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BLCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| BRCA | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| CESC | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| COAD | |||||||
| ESCA | |||||||
| ESCA | |||||||
| GBM | |||||||
| HNSC | |||||||
| HNSC | |||||||
| HNSC | |||||||
| KIRC | |||||||
| KIRP | |||||||
| LGG | |||||||
| LGG | |||||||
| LGG | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LIHC | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUAD | |||||||
| LUSC | |||||||
| LUSC | |||||||
| LUSC | |||||||
| OV | |||||||
| OV | |||||||
| OV | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PAAD | |||||||
| PRAD | |||||||
| PRAD | |||||||
| READ | |||||||
| READ | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SARC | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| SKCM | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| STAD | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC | |||||||
| UCEC |
| Cancer | Type | Freq | Q-value |
|---|---|---|---|
| CHOL | |||
| LAML | |||
| LGG | |||
| LIHC | |||
| PRAD | |||
| THCA | |||
| THYM | |||
| UCS |
| Cancer | P-value | Q-value |
|---|---|---|
| THYM | ||
| KIRC | ||
| ACC | ||
| UCS | ||
| HNSC | ||
| PRAD | ||
| LUSC | ||
| BRCA | ||
| PAAD | ||
| BLCA | ||
| READ | ||
| UCEC | ||
| LIHC | ||
| LGG | ||
| CHOL | ||
| LUAD | ||
| OV |
| Transcript ID | Name | Length | RefSeq ID | Protein ID | Length | RefSeq ID | UniportKB ID |
|---|---|---|---|---|---|---|---|
| ENST00000627871 | STXBP1-210 | 538 | - | ENSP00000485895 | 100 (aa) | - | A0A0D9SEP9 |
| ENST00000373299 | STXBP1-201 | 3765 | - | ENSP00000362396 | 594 (aa) | - | P61764 |
| ENST00000636962 | STXBP1-216 | 2918 | - | ENSP00000489762 | 574 (aa) | - | A0A1B0GWF2 |
| ENST00000637173 | STXBP1-218 | 4193 | - | ENSP00000490519 | 580 (aa) | - | A0A0D9SG72 |
| ENST00000496504 | STXBP1-205 | 591 | - | ENSP00000485361 | 75 (aa) | - | A0A096LP33 |
| ENST00000626539 | STXBP1-209 | 3840 | - | ENSP00000487211 | 580 (aa) | - | A0A0D9SG72 |
| ENST00000637521 | STXBP1-220 | 2068 | - | ENSP00000489791 | 589 (aa) | - | A0A1B0GTP9 |
| ENST00000637464 | STXBP1-219 | 4893 | - | ENSP00000489655 | 55 (aa) | - | A0A1B0GTD8 |
| ENST00000494254 | STXBP1-204 | 563 | - | ENSP00000485397 | 119 (aa) | - | A0A096LP52 |
| ENST00000635950 | STXBP1-214 | 2771 | - | ENSP00000490903 | 574 (aa) | - | A0A1B0GWF2 |
| ENST00000626333 | STXBP1-207 | 667 | - | ENSP00000486814 | 147 (aa) | - | A0A0D9SFQ7 |
| ENST00000647107 | STXBP1-222 | 1843 | - | ENSP00000494727 | 580 (aa) | - | A0A2R8Y5D4 |
| ENST00000626416 | STXBP1-208 | 3700 | - | - | - (aa) | - | - |
| ENST00000637060 | STXBP1-217 | 3825 | - | ENSP00000490674 | 112 (aa) | - | A0A1B0GVV9 |
| ENST00000625363 | STXBP1-206 | 550 | - | ENSP00000486944 | 149 (aa) | - | A0A0D9SFW6 |
| ENST00000636509 | STXBP1-215 | 3894 | - | ENSP00000490810 | 360 (aa) | - | A0A1B0GW76 |
| ENST00000628638 | STXBP1-211 | 581 | - | - | - (aa) | - | - |
| ENST00000373302 | STXBP1-202 | 3967 | - | ENSP00000362399 | 603 (aa) | - | P61764 |
| ENST00000628768 | STXBP1-212 | 2582 | - | - | - (aa) | - | - |
| ENST00000630492 | STXBP1-213 | 572 | - | ENSP00000485680 | 102 (aa) | - | A0A0D9SEH5 |
| ENST00000476182 | STXBP1-203 | 566 | - | ENSP00000490199 | 22 (aa) | - | A0A1B0GUQ2 |
| ENST00000637953 | STXBP1-221 | 3925 | - | ENSP00000490613 | 577 (aa) | - | A0A1B0GVQ5 |
| Pathway ID | Pathway Name | Source |
|---|---|---|
| hsa04721 | Synaptic vesicle cycle | KEGG |
| ensgID | SNP | Chromosome | Position | SNP-risk | Trait | PubmedID | 95% CI | Or or BEAT | EFO ID |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000136854 | rs10819303 | 9 | 127630675 | A | Educational attainment (MTAG) | 30038396 | [0.0056-0.0106] unit decrease | 0.0081 | EFO_0004784 |
| ENSG00000136854 | rs10819303 | 9 | 127630675 | A | Highest math class taken (MTAG) | 30038396 | [0.0063-0.0129] unit decrease | 0.0096 | EFO_0004875 |
| ENSG00000136854 | rs2039204 | 9 | 127608828 | A | Educational attainment (years of education) | 30038396 | [0.0055-0.0109] unit increase | 0.0082 | EFO_0004784 |
| ENSG00000136854 | rs1573178 | 9 | 127610208 | T | Educational attainment (years of education) | 30038396 | [0.006-0.0114] unit decrease | 0.0087 | EFO_0004784 |
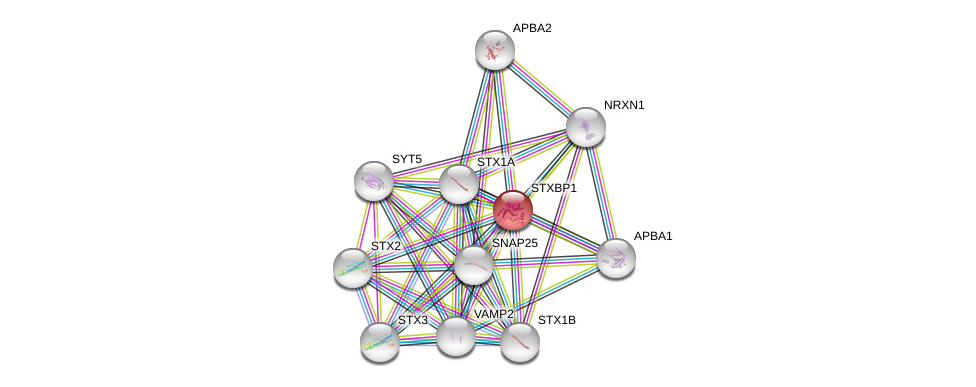
| Go ID | Go_term | PubmedID | Evidence | Category |
|---|---|---|---|---|
| GO:0000149 | SNARE binding | - | ISS | Function |
| GO:0002576 | platelet degranulation | 12773094. | IMP | Process |
| GO:0003006 | developmental process involved in reproduction | - | IEA | Process |
| GO:0003723 | RNA binding | 22658674. | HDA | Function |
| GO:0005515 | protein binding | 25416956.25910212. | IPI | Function |
| GO:0005654 | nucleoplasm | - | IDA | Component |
| GO:0005737 | cytoplasm | - | ISS | Component |
| GO:0005739 | mitochondrion | - | ISS | Component |
| GO:0005829 | cytosol | - | IDA | Component |
| GO:0005829 | cytosol | - | TAS | Component |
| GO:0005856 | cytoskeleton | - | IEA | Component |
| GO:0005886 | plasma membrane | 12773094. | IDA | Component |
| GO:0005886 | plasma membrane | - | ISS | Component |
| GO:0006904 | vesicle docking involved in exocytosis | - | ISS | Process |
| GO:0007269 | neurotransmitter secretion | - | TAS | Process |
| GO:0007274 | neuromuscular synaptic transmission | - | IEA | Process |
| GO:0007412 | axon target recognition | - | ISS | Process |
| GO:0010807 | regulation of synaptic vesicle priming | - | ISS | Process |
| GO:0014047 | glutamate secretion | - | TAS | Process |
| GO:0015031 | protein transport | - | IEA | Process |
| GO:0016082 | synaptic vesicle priming | - | IEA | Process |
| GO:0016188 | synaptic vesicle maturation | - | ISS | Process |
| GO:0017075 | syntaxin-1 binding | 12730201.19748891. | IPI | Function |
| GO:0017075 | syntaxin-1 binding | - | ISS | Function |
| GO:0019901 | protein kinase binding | - | IEA | Function |
| GO:0019904 | protein domain specific binding | - | IEA | Function |
| GO:0019905 | syntaxin binding | 12773094. | IPI | Function |
| GO:0030424 | axon | - | IEA | Component |
| GO:0031091 | platelet alpha granule | 12773094. | IDA | Component |
| GO:0031333 | negative regulation of protein complex assembly | - | IEA | Process |
| GO:0031630 | regulation of synaptic vesicle fusion to presynaptic active zone membrane | 15217342. | TAS | Process |
| GO:0032229 | negative regulation of synaptic transmission, GABAergic | - | ISS | Process |
| GO:0032355 | response to estradiol | - | IEA | Process |
| GO:0032991 | protein-containing complex | - | ISS | Component |
| GO:0035542 | regulation of SNARE complex assembly | 15217342. | TAS | Process |
| GO:0042802 | identical protein binding | - | ISS | Function |
| GO:0043274 | phospholipase binding | - | IEA | Function |
| GO:0043306 | positive regulation of mast cell degranulation | - | IEA | Process |
| GO:0043524 | negative regulation of neuron apoptotic process | - | IEA | Process |
| GO:0045335 | phagocytic vesicle | - | IEA | Component |
| GO:0045956 | positive regulation of calcium ion-dependent exocytosis | - | IEA | Process |
| GO:0047485 | protein N-terminus binding | - | IEA | Function |
| GO:0048471 | perinuclear region of cytoplasm | - | IEA | Component |
| GO:0048787 | presynaptic active zone membrane | - | IEA | Component |
| GO:0050821 | protein stabilization | - | IEA | Process |
| GO:0060292 | long-term synaptic depression | - | IEA | Process |
| GO:0070062 | extracellular exosome | 19056867. | HDA | Component |
| GO:0070527 | platelet aggregation | 12773094. | IMP | Process |
| GO:0071346 | cellular response to interferon-gamma | - | IEA | Process |
| GO:0072659 | protein localization to plasma membrane | 17543282. | IDA | Process |
| GO:0098794 | postsynapse | - | IEA | Component |
| GO:0098978 | glutamatergic synapse | - | IEA | Component |
| GO:0099525 | presynaptic dense core vesicle exocytosis | - | IEA | Process |
| GO:0106022 | positive regulation of vesicle docking | - | IEA | Process |
| GO:1903296 | positive regulation of glutamate secretion, neurotransmission | - | IEA | Process |